Open Access Article
Open Access ArticleSelf-assembled Pt2L2 boxes strongly bind G-quadruplex DNA and influence gene expression in cancer cells†
O.
Domarco
a,
D.
Lötsch
b,
J.
Schreiber
b,
C.
Dinhof
b,
S.
Van Schoonhoven
b,
M. D.
García
a,
C.
Peinador
*a,
B. K.
Keppler
cd,
W.
Berger
bd and
A.
Terenzi
 *cd
*cd
aUniversidade da Coruña, Departamento de Química Fundamental and Centro de Investigacións Científicas Avanzadas, Facultade de Ciencias, E-15071 A Coruña, Spain. E-mail: carlos.peinador@udc.es
bMedical University Vienna, Department of Medicine I, Institute of Cancer Research and Comprehensive Cancer Center, Borschkegasse 8a, A-1090 Vienna, Austria
cUniversity of Vienna, Institute of Inorganic Chemistry, Waehringerstrasse 42, A-1090 Vienna, Austria
dResearch Platform “Translational Cancer Therapy Research”, University of Vienna and Medical University of Vienna, Vienna, Austria. E-mail: alessio.terenzi@univie.ac.at
First published on 23rd November 2016
Abstract
Supramolecular Pt(II) quadrangular boxes bind native and G-quadruplex DNA motifs in a size-dependent fashion. Three Pt molecular squares of distinct size show biological activity against cancer cells and heavily influence the expression of genes known to form G-quadruplexes in their promoter regions. The smallest Pt-box displays less activity but more selectivity for a quadruplex formed in the c-Kit gene.
There is considerable interest in the development of metallo-supramolecular DNA-binders.1 They are species able to interact through non-covalent interactions with nucleic acids and, in principle, to discriminate between different DNA tertiary structures. Among these, G-quadruplexes (G4s), G-rich sequences able to form four-stranded structures organized in stacked guanine tetrads,2 are appealing targets for anticancer drugs, as they are overrepresented in telomeres and oncogenes with important regulatory functions.2–4
In contraposition with traditional covalent synthetic chemistry, coordination-driven self-assembly allows for the efficient synthesis of nanometric aggregates, affording supramolecules that can potentially mimic the dimensions and binding capacities of proteins with DNA recognition motifs (e.g. helicases).1c,4 In this context, since the seminal work by the groups of Fujita and Stang,5 Pt(II)-directed self-assembly has enabled the modular preparation of innumerable supramolecular structures.6 Surprisingly, after the discovery that Fujita's Pt(II) square [Pt(en)(4,4′-dipyridyl)]4 was an effective telomeric G4 binder, only a small number of studies have been reported on the interesting G4 binding capability of square and rectangular-shaped self-assembled Pt(II) metallacycles.8–10
In recent years, some of us have developed a self-assembly strategy for the synthesis of molecular rectangles and squares based on N-monoalkylated 4,4′-bipyridine/diazapyrene ligands and Pt(II)/Pd(II) metal centres.11 We found that a Pt(II) dinuclear rectangle based on a 2,7-diazapyrenium ligand binds and bends native DNA, and interferes with DNA transactions in vitro, resulting in a compound which shows similar levels of efficacy, but a different spectrum of activity, to cisplatin.12
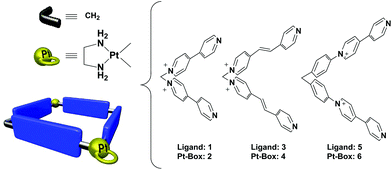
With the aim to demonstrate that our self-assembly strategy can be exploited for the fast and modular generation of libraries of Pt(II) metallacycles, with improved binding affinity and selectivity towards specific DNA topologies, we herein present the evaluation as DNA binders of three Pt(II) dinuclear squares of different sizes 2, 4 and 6 and their constituent L-shaped 4,4′-bipyridine ligands 1, 3 and 5 (Fig. 1). Strategically, we selected Pt-boxes having slightly increasing dimensions (2: 110 Å2, 4: 162 Å2, 6: 220 Å2), compared to the area of the G-quartet ∼92 Å2.‡ In the present communication, we show our findings on the interaction of metallacycles and their ligands with double stranded DNA and selected G4s from human telomere and gene promoter regions. Moreover, we discuss the effects of these compounds on cancer cell lines, considering their gene expression regulation.
Ligands 1, 3 and 5, and the corresponding Pt-boxes 2, 4 and 6, were prepared according to previously reported methodologies,13 and used as their water soluble nitrate salts.
To test the binding ability of ligands and Pt-boxes towards the selected DNA motifs, we performed FRET melting assays, using porphyrin TMPyP4 as a non-selective binder control, both for duplex and G4s.14,15 FRET melting profiles of 0.2 μM oligonucleotides upon the interaction of 1 μM binders (Fig. 2 and S1, ESI†) show a minor/null activity of the ligands and, in contrast, a remarkable interaction of the Pt complexes. Pt-Boxes 4 and 6, in particular, demonstrate to strongly interact with G4s and ds-DNA with ΔT1/2 values even higher than the control (Table 1).
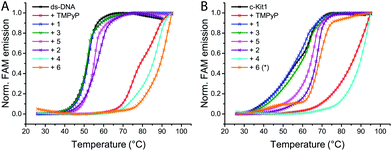 | ||
| Fig. 2 Representative FRET melting profiles of ds-DNA (A) and the G-quadruplex c-Kit1 (B), upon interaction of compounds 1–6 and control TMPyP4. Buffer: 60 mM potassium cacodylate, pH 7.4. Melting profiles for all compounds in the ESI.† *Compound 6 concentration = 0.25 μM. | ||
| TMPyP | 1 | 3 | 5 | 2 | 4 | 6 | |
|---|---|---|---|---|---|---|---|
| a Compound 6 concentration = 0.25 μM. | |||||||
| ds-DNA | 27.2 | 0.2 | −0.4 | 3.5 | 4.9 | 33.7 | 37.5 |
| h-Telo | 31.8 | −1.5 | −0.4 | 3.9 | 1.5 | 36.9 | 12.3a |
| c-Myc | 11.8 | −2.9 | −2.5 | 0.8 | 1.7 | 19.2 | 1.7a |
| c-Kit1 | 28.7 | −0.7 | 2.3 | 7.6 | 10.2 | 32.8 | 10.2a |
| h-TERT | 26.4 | 0.9 | 2.1 | 3.5 | 5.4 | 23.8 | 30.1 |
| bcl2 | 35.0 | −2.7 | −1.8 | 0.8 | 3.8 | 32.2 | 36.2 |
| TERRA | 14.9 | −0.9 | 0.6 | 3.8 | 4.0 | 19.9 | 4.0a |
For compound 6, in most of the cases, we had to reduce its concentration down to 0.25 μM in order to see an appreciable FAM emission, since at 1 μM it was not possible to “melt” the structures (Fig. S2, ESI†). These results highlight that the Pt-box bigger in size (6) is the best DNA binder regardless of the DNA tertiary structure. Surprisingly, the smaller compound 2 shows a modest interaction with ds-DNA while it is able to significantly bind c-Kit1 G4 and, to less extent, h-Tert G4 (Table 1).
This last result emphasises the few hints found in the literature suggesting that modulating the size of Pt-squares can result in geometries for effective targeting of a specific G4 structure.7–9 In this direction, molecular docking studies (Fig. S3, ESI†) show our Pt-boxes 2, 4 and 6 placing themselves on the top/end faces of the telomeric quadruplex (h-Telo), as already found for [Pt(en)(4,4′-dipyridyl)]4.7 On the other hand, they bind in different sites to the more heterogeneous c-Kit1 G4. Interestingly, compound 2 behaves differently compared to the other boxes in both G4s, being the only one fitting into the unique binding pocket of c-Kit1 G4.16
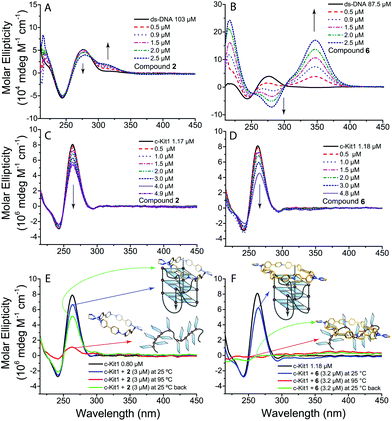
To obtain more information on the interaction of metallacycles 2, 4 and 6 with ds-DNA and c-Kit1 G4 we performed circular dichroism (CD) and UV-Vis titrations. The CD spectra in Fig. 3B confirm the strong interaction of compound 6 with ds-DNA, with the appearance of a distinctive induced CD band (ICD) at ≈350 nm, together with heavy modification of ds-DNA characteristic CD bands (black line), indicating a possible bending effect.12 Pt-Box 2 interaction with ds-DNA is by far weaker (Fig. 3A), producing just a small ICD band with the B-DNA structure being retained. Furthermore, as shown in Fig. 3C/D, the intensity of the c-Kit1 band at ≈260 nm is lowered upon interaction with 2 and 6, with 6 > 2. Overall, the CD effects produced by titrating ds-DNA and c-Kit1 G4 with the Pt-boxes increase with the compound size (6 >4 > 2, for all CD titrations see Fig. S4–S9, ESI†).
We monitored the interaction with c-Kit1 by variable temperature CD to see the effect of the Pt-boxes on the G4 folding process. The c-Kit1 G4 structure is retained in solution after a melting–annealing cycle with compound 2 but, surprisingly, is not able to form anymore after the interaction with 6 (Fig. 3E and F). The latter interacts so strongly even with the non-folded form at 95 °C that, once the temperature is decreased back at 25 °C, the oligonucleotide is not capable of rearranging itself into a G4 fashion. This is another hint of the importance of the Pt-box size on the binding affinity. Pt-Box 4 shows the same behaviour as 6, although slightly less pronounced (Fig. S10, ESI†). Kb values, obtained from UV-Vis titrations (Fig. S11–S14†), indicate the tight binding of compounds 4 and 6 with ds-DNA with Kb values of (1.3 ± 0.2) × 105 and (2.3 ± 0.4) × 105 M−1, respectively, and increased affinity for c-Kit1 with Kb values of (3.5 ± 1.0) × 106 and (2.0 ± 0.3) × 107 M−1, respectively.
With the above commented cell-free encouraging results on the interaction of the Pt-boxes 2, 4 and 6 with DNA and G4s, we decided to determine their effect on cancer cells, in comparison with their constituent 4,4′-bipyridine-based ligands 1, 3 and 5. Therefore, three cancer cell lines (osteosarcoma U2OS, melanoma VM-1 and breast cancer MCF-7) were treated for 72 hours with increasing drug concentrations and the MTT assay was performed (Table 2).
| Cisplatin | 1 | 3 | 5 | 2 | 4 | 6 | |
|---|---|---|---|---|---|---|---|
| U2OS | 3.8 ± 3.0 | >50 | 38.2 ± 3.4 | >50 | 37.8 ± 3.3 | 28.9 ± 4.1 | 42.3 ± 4.6 |
| VM-1 | 4.43 ± 0.05 | >50 | >50 | >50 | >50 | 31.8 ± 0.5 | >50 |
| MCF7 | 6.7 ± 0.3 | >50 | >50 | >50 | 32.1 ± 7.4 | 39.4 ± 3.4 | 39.6 ± 4.5 |
While the ligands 1, 3 and 5 are widely inactive (with the exception of 3 showing some activity on U2OS), the Pt compounds (2, 4 and 6) reduce cell viability in the majority of the tested cell models, with IC50 values ranging from ∼32 to 45 μM. Interestingly, the middle-sized 4 is the most active against U2OS, being the only Pt-box active against the strongly cisplatin resistant cell line VM-1. We investigated the impact of ligands and Pt-boxes on the cell cycle progression (Fig. S15, ESI†), observing that 48-hour exposure to the ligands (1, 3 and 5) does not exert pronounced effects in any tested cancer cell line. Interestingly, Pt compound 6 (25 μM) produces a significant G0–G1 arrest in MCF-7 cells, whilst it generates a distinct G2-M phase increase in the melanoma model; both results consistent with DNA damage mechanisms. After the treatment with Pt-box 4, an induction of cell death is observed in melanoma VM-1 cells, indicated by an increase in the amount of cells localizing on the sub-G0–G1 fraction.
To confirm the differences of the investigated compounds in their apoptosis-mediating activity, cell surface exposure of phosphatidylserine was tested by Annexin V labelling and FACS analysis after 24 h drug exposure at 25 μM. Co-staining with propidium iodide (PI) indicates dead cells either from necrosis (Annexin −, PI +) or late apoptosis (Annexin +, PI +) (Fig. S16, ESI†). Concerning the 4,4′-bipyridine-based ligands, only 3 induces a minor increase of apoptotic cells in the breast cancer and melanoma cell models. While the platinum compounds 2 and 6 result in low to moderately enhanced apoptosis levels, the middle-sized Pt-box 4 exerts induction of both apoptotic and necrotic cell death.
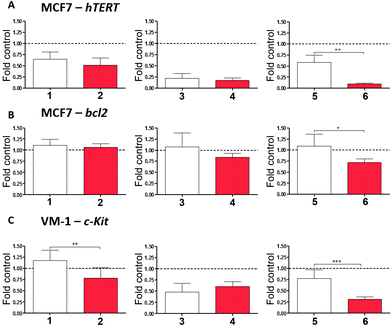
All cell-free experiments confirmed that the tested Pt(II) compounds, but not their ligands, interact with ds-DNA, as well as with the G4 of c-Kit1, hTERT and bcl2 gene promoters (see Table 1). In order to investigate a possible impact of the Pt-boxes on the mRNA level of these potential target genes, we treated the cancer cells with our compounds, extracted the RNA and performed real-time PCR analyses (see Methods in the ESI†). hTERT and bcl2 were analysed in MCF-7 breast cancer cells, while c-Kit was evaluated on VM-1 melanoma cells, as MCF-7 cells lacked detectable c-Kit1 mRNA expression (Fig. 4).
As a general trend, the final output of the experiment is that the bigger the Pt-box, the higher is the downregulation of the selected genes (Fig. 4), notably corroborating the cell-free tests.
When compared to the corresponding ligands as internal controls, the bigger-sized compound 6 significantly reduces the expression levels of the three genes upon 24 hours of exposure, being particularly active on the expression of hTERT. The middle-sized Pt-box 4 reduces the expression levels of hTERT and c-Kit, but also its ligand 3 produces the same results, indicating, together with the MTT assay and cell cycle results, a possible difference in the mechanism of action for the Pt-box bearing a double bond in the 4,4′-bipyridine-based scaffold. The smaller compound 2 produces a selective effect on the expression of c-Kit, its ligand being not active. This latter result is in good agreement with the spectroscopic findings, as well as with the molecular modelling studies, suggesting that the smaller Pt-box has less affinity but better selectivity for a particular G4 topology.
In conclusion, we proved that it is possible to modulate the DNA binding activity and the in vitro effect on cancer cells with Pt supramolecular architectures according to their sizes and shapes. We are currently using our self-assembly strategy to build a library of novel Pt compounds for the fine-tuning of affinity and selectivity towards specific G-quadruplex structures.
A. T. has received funding from the Mahlke-Obermann Stiftung and the European Union's Seventh Framework Programme (grant agreement no. 609431). O. D., M. D. G. and C. P. also thank Ministerio de Economía y Competitividad (MINECO FEDER, CTQ201341097-P) and Xunta de Galicia (EM2014/056) for financial support.
Notes and references
- For selected reviews, see: (a) B. J. Pages, D. L. Ang, E. P. Wright and J. R. Aldrich-Wright, Dalton Trans., 2015, 44, 3505 RSC; (b) S. N. Georgiades, N. H. Abd Karim, K. Suntharalingam and R. Vilar, Angew. Chem., Int. Ed., 2010, 49, 4020 CrossRef CAS PubMed; (c) M. J. Hannon, Chem. Soc. Rev., 2007, 36, 280 RSC.
- (a) D. Rhodes and H. J. Lipps, Nucleic Acids Res., 2015, 43, 8627 CrossRef CAS PubMed; (b) S. Balasubramanian, L. H. Hurley and S. Neidle, Nat. Rev. Drug Discovery, 2011, 10, 261 CrossRef CAS PubMed.
- S. Neidle, J. Med. Chem., 2016, 59, 5987 CrossRef CAS PubMed.
- O. Mendoza, A. Bourdoncle, J. B. Boulé, R. M. Brosh and J. L. Mergny, Nucleic Acids Res., 2016, 44, 1989 CrossRef CAS PubMed.
- (a) M. Fujita, Chem. Soc. Rev., 1998, 27, 417 RSC; (b) P. J. Stang and B. Olenyuk, Acc. Chem. Res., 1997, 30, 502 CrossRef CAS.
- For selected recent reviews, see: (a) R. Chakrabarty, P. S. Mukherjee and P. J. Stang, Chem. Rev., 2011, 111, 6810 CrossRef CAS PubMed; (b) T. R. Cook, Y. R. Zheng and P. J. Stang, Chem. Rev., 2013, 113, 734 CrossRef CAS PubMed; (c) A. Mishra, S. C. Kang and K. W. Chi, Eur. J. Inorg. Chem., 2013, 30, 5222 CrossRef; (d) S. Mukherjee and P. S. Mukherjee, Chem. Commun., 2014, 50, 2239 RSC; (e) T. R. Cook and P. J. Stang, Chem. Rev., 2015, 115, 7001 CrossRef CAS PubMed.
- R. Kieltyka, P. Englebienne, J. Fakhoury, C. Autexier, N. Moitessier and H. F. Sleiman, J. Am. Chem. Soc., 2008, 130, 10040 CrossRef CAS PubMed.
- X. H. Zheng, H. Y. Chen, M. L. Tong, L. N. Ji and Z. W. Mao, Chem. Commun., 2012, 48, 7607 RSC.
- X. H. Zheng, Y. F. Zhong, C. P. Tan, L. N. Ji and Z. W. Mao, Dalton Trans., 2012, 41, 11807 RSC.
- S. Ghosh, O. Mendoza, L. Cubo, F. Rosu, V. Gabelica, A. J. P. White and R. Vilar, Chem. – Eur. J., 2014, 20, 4772 CrossRef CAS PubMed.
- (a) M. D. García, C. Alvariño, E. M. López-Vidal, T. Rama, C. Peinador and J. M. Quintela, Inorg. Chim. Acta, 2014, 417, 27 CrossRef; (b) C. Peinador, V. Blanco, M. García and J. Quintela, in Molecular Self-Assembly, Pan Stanford Publishing, 2012, pp. 351–382 Search PubMed.
- A. Terenzi, C. Ducani, V. Blanco, L. Zerzankova, A. F. Westendorf, C. Peinador, J. M. Quintela, P. J. Bednarski, G. Barone and M. J. Hannon, Chem. – Eur. J., 2012, 18, 10983 CrossRef CAS PubMed.
- (a) Ligand 1 and Pt box 2: V. Blanco, M. Chas, D. Abella, E. Pía, C. Platas-Iglesias, C. Peinador and J. M. Quintela, Org. Lett., 2008, 10, 409 CrossRef CAS PubMed; (b) Ligand 5 and Pt-box 6: V. Blanco, A. Gutiérrez, C. Platas-Iglesias, C. Peinador and J. M. Quintela, J. Org. Chem., 2009, 74, 6577 CrossRef CAS PubMed; (c) Ligand 3 and Pt-box 4: E. M. López-Vidal, V. Blanco, M. D. García, C. Peinador and J. M. Quintela, Org. Lett., 2012, 14, 580 CrossRef PubMed.
- A. De Cian, L. Guittat, M. Kaiser, B. Saccà, S. Amrane, A. Bourdoncle, P. Alberti, M. P. Teulade-Fichou, L. Lacroix and J. L. Mergny, Methods, 2007, 42, 183 CrossRef CAS PubMed.
- F. X. Han, R. T. Wheelhouse and L. H. Hurley, J. Am. Chem. Soc., 1999, 121, 3561 CrossRef CAS.
- A. T. Phan, V. Kuryavyi, S. Burge, S. Neidle and D. J. Patel, J. Am. Chem. Soc., 2007, 129, 4386 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental methods, and additional spectroscopic and docking data/pictures. See DOI: 10.1039/c6dt03876j |
| ‡ The G-quadruplex area was calculated by measuring the distances of the two sides of the G-quartet, selecting the guanine nitrogen atoms attached to the phosphate backbone. We used the “distance tool” in Chimera software and the final area is the average of four G4s (PDB IDs: 1KF1, 2O3M, 1XAV and 2F8U). |
| This journal is © The Royal Society of Chemistry 2017 |