Recent progress in surface-enhanced Raman spectroscopy for biological and biomedical applications: from cells to clinics
D.
Cialla-May†
abc,
X.-S.
Zheng†
 b,
K.
Weber
abc and
J.
Popp
b,
K.
Weber
abc and
J.
Popp
 *abc
*abc
aFriedrich Schiller University Jena, Institute of Physical Chemical and Abbe Center of Photonics, Helmholtzweg 4, 07745 Jena, Germany. E-mail: juergen.popp@uni-jena.de
bLeibniz Institute of Photonic Technology Jena, Albert-Einstein-Str. 9, 07745 Jena, Germany
cResearch Campus Infectognostic, Philosophenweg 7, 07743 Jena, Germany
First published on 22nd June 2017
Abstract
The application of surface-enhanced Raman spectroscopy (SERS) in biological and biomedical detection schemes is feasible due to its excellent molecular specificity and high sensitivity as well as the capability of SERS to be performed in complex biological compositions. SERS-based investigation of cells, which are the basic structure and functional unit of organisms, represents the starting point of this review. It is demonstrated that SERS provides a deep understanding of living cells as well as their microenvironment which is needed to assess the development of diseases. The clinical relevance of SERS is proved by its application for the detection of cancer cells and tumour margins under in vivo conditions and examples for theranostic approaches are discussed. This review article provides a comprehensive overview of the recent progress within the last 3 years.
1 Introduction
An understanding of biological processes is of great importance for the detection of diseases and fast initiation of therapeutic measures. Moreover, specific and reliable analysis of metabolites is needed for gaining deep insight into cellular processes or the assessment of therapeutic success. Therefore, methods are required allowing for fast, molecular specific and trace sensitive detection in complex biological environments. The application of surface-enhanced Raman spectroscopy (SERS) for biological and medical applications gained much attention in the last decade which is attributed to the molecular specificity of Raman spectroscopy combined with high sensitivity due to the optical properties of plasmonic nanostructures. SERS is applied to characterize the interaction of biomolecules with metallic surfaces, to quantify drugs and biomolecules in complex matrices, to investigate cells and tissues related to e.g. cancer diagnostics and even in vivo detection schemes are presented employing this method.1–3Based on the interaction of electromagnetic waves with metallic nanoparticles with dimensions much smaller than the wavelength, a displacement of the electron cloud against the atomic cores is caused and, thus, a polarization within the metal nanoparticle is induced. This resonant excitation due to the resonant electron dynamic and related local field enhancement is referred to as a localized surface plasmon polariton (LSPP). The spectral position of the LSP resonance is defined by the plasmon resonance condition and is a function of the dielectric constant of the surrounding medium, and the size, shape and material of the metal nanoparticle.4 Additionally, propagating surface plasmon polariton (PSPP) modes occur at the interface between a metallic waveguide and a dielectric and, in the case of a periodic nanostructured surface, its frequency is tunable by the angle of incidence of the light as well as lattice parameters such as period.5 The excitation of surface plasmon polarition modes which are associated with a strong field enhancement is known as the electromagnetic mechanism, the main contribution to SERS enhancement.6 Theoretical calculations demonstrate that the maximum electromagnetic enhancement factor is around 1011; therefore, the molecule needs to be located in close vicinity to the metallic surface at hot spots. The second contribution to the overall SERS response is the chemical enhancement, due to the chemical interaction between the molecule and the nanostructured surface in the ground state. This chemical interaction results in the formation of a charge-transfer state between the nanoparticle and the molecule. This charge transfer state can be resonantly excited leading to a resonance Raman enhancement. The chemical enhancement factor is in the range of 101 and 103.
For the interpretation of SERS spectra, the orientation of the molecule of interest towards the metal surface needs to be considered due to its influence on the spectral shape of SERS spectra. This effect is described as surface selection rules, meaning that Raman modes perpendicular to the metal surface are preferentially enhanced and, thus, dominate the SERS spectrum. Further on, due to the evanescent character of the electromagnetic field on the metal surface, only Raman modes in close vicinity (up to 10 nm) can contribute to the overall SERS spectrum, which is important for the spectroscopic investigations of large bio- and macromolecules. The binding toward the metallic surface is preferably via sulfur, nitrogen and oxygen containing groups.2
In general, two types of methodologies are available for SERS-based biological and biomedical applications i.e. label-free detection as well as indirect approaches applying SERS tags. SERS-based label-free detection is to obtain vibrational spectroscopic information of biomolecules via the direct interaction between the samples and the SERS-based nanostructures, thus, providing intrinsic fingerprint information of biological and biomedical samples with enhanced intensity. Indirect detection usually utilizes Raman reporter molecules with strong and distinct Raman signals to label the SERS tags, which can be used for ultra-sensitive SERS-based detection, imaging, or sensing of biological and biomedical samples. By taking advantage of an additional resonance Raman enhancement, surface enhanced resonance Raman spectroscopy (SERRS) is created, which is associated with the application of dyes as reporter molecules for SERS tags. Thus, the overall spectral response is dominated by the contribution of the dye and the spectra are easier to interpret in comparison to label-free or direct SERS approaches. Powerful and rational design of plasmonic nanostructures for SERS is essential for the successful application of both label-free SERS and indirect SERS in biological and biomedical analysis. Since various SERS-based nanostructures are involved in many detection schemes for biological and biomedical analysis, an overview of the plasmonic nanostructures for SERS will be given first.
Generally, nanostructures for SERS-based biological and biomedical applications consist of four parts: (1) a substrate of noble metallic nanostructure showing strong SERS activity, which is based mainly on silver or gold and is prepared by bottom up, self-organization and top-down processes; (2) a layer of reporter molecules with a unique and strong Raman fingerprint enabling indirect detection; (3) an outer coating layer to improve biocompatibility and stability; and (4) bio-conjugations allowing for targeting and specific detection. Not all the above four parts are necessary for the design of nanostructures for SERS, especially for label-free SERS detection. Here, bare nanostructures are usually applied to directly interact with biological samples to obtain the vibrational information of the biological samples. However, for both label-free and indirect detection, the noble metallic nanostructure is essential since the detection sensitivity directly relies on its structure and optical properties. An effective enhancement can be achieved for various excitation wavelengths by tuning the LSP resonance due to changes in size, shape or coupling modes. For in vivo studies and photothermotherapy, structures with strong absorption in the near infrared (NIR) region are desirable. In indirect SERS detection, additionally to provide a unique Raman fingerprint for labelling and imaging, the reporter molecule layer can be further modified to be sensitive to the microenvironments. In the case of indirect detection of living cells and in vivo systems, the outer encapsulation layer is necessary, which can not only ensure biocompatibility, but also improve stability by avoiding desorption of the reporter molecules from the metal surface and adsorption of unwanted molecules in the detection system to the metal surface. Available encapsulation materials used in the literature are proteins, DNA, polymers and silica. Finally, further bio-conjugations with targeting peptides, antibodies, or aptamers are also alternatives for targeting and specific detection. This short summary provides the fundamental instructions for SERS-based nanostructures for biological and biomedical applications; however, the readers could find more detailed information in the following sections together with the highlights of recent progress in SERS-based biological and biomedical detection or may refer to review articles published on this particular topic.3,6
In this review, SERS as a powerful analytical tool in biology and medicine is discussed. First, the characterization of living cells and the investigation of the cellular microenvironment are summarized in sections 2 and 3, respectively. Moreover, section 4 gives an overview about the detection of cancer and circulating tumour cells (CTCs), and the identification of tumour margins, and examples for in vivo applications of SERS in medicine are highlighted. Finally, the theranostic capabilities of SERS are shown and comprehensively discussed in section 5. The progress and perspective are summarized in section 6. Thus, this review article gives detailed information about selected facets SERS has when it comes to an application in biology and medicine.
2 SERS-based studies on living cells
Cells are the basic structural and functional unit of living organisms. It is a dynamic and heterogeneous system undergoing rapid changes and constant matter and energy exchange with the environment, which results in great difference in life activities of cells, both in time and space. Intercellular signalling, which is the basis for the realization of various biological functions and life activities (such as cell apoptosis, proliferation, differentiation and changes in cell components or its microenvironments), is intimately associated with the occurrence and progression of diseases. As a result, investigation of cells has long been one of the most fundamental and important areas in biomedical research. Conventional biomedical techniques, e.g. cell staining and tissue sectioning, do not meet the requirements of further investigations of biological systems and the desire for a better understanding of diseases. Instead, increasing attention has been paid to exploring diseases at the single cell and single molecule level. SERS, combining nanotechnology with molecular spectroscopy and holding the advantages of high sensitivity, high spectral resolution, reliable signals free from quenching or photobleaching, non-invasive measurements with minimum damage to biological samples, as well as little interference from autofluorescence and water, has been a powerful tool widely used for various studies of living cells. In this section, we review the recent progress regarding direct intracellular analysis, investigation of biological processes and cellular functions, as well as SERS imaging of cell-surface species.2.1 Direct intracellular analysis
In this section, we discuss the intrinsic analysis of cellular information by using label-free SERS-based detection. With this method, intrinsic spectroscopic signatures from living cells can be acquired to identify different cell components.The application of SERS in the characterization of fundamental cellular processes is limited due to the challenge of selectively identifying molecular changes from cellular processes in large and multidimensional data sets and the lack of simple tools for extracting this information. To overcome this limitation, a reference-based PCA–LDA score differential analysis methodology was developed for the extraction of unknown features/spectra from a hyperspectral data set and the subsequent generation of pseudocolor SERS maps and biochemical SERS characterization.7 As illustrated in Fig. 1, the SERS spectra can be segregated from the endosomal and lysosomal pathways within the endocytic uptake system by combining this method with an experimental gold nanoparticle (AuNP) pulse-depletion approach. Subsequently, the results were used to generate pseudocolor maps indicating the distribution of endosomes and lysosomes via chemometric methods. This approach enables the application of label-free SERS to identify unknown components in living cells. In addition, 3D SERS imaging of intracellular pathways with AuNPs was also realized.8 Dynamic SERS imaging with 3D mapping in a living cell by combining a confocal Raman spectrometer and a dual-focus dark-field microscope was demonstrated. AuNPs travel through the living cell acting as a probe. They are tracked by dual-focus dark-field imaging with position feedback to the confocal Raman spectrometer in order to record evolving SERS signals from the same gold nanoparticle moving through the cell. Simultaneous detection of motion and recording of the SERS spectrum enable the visualization of the SERS pathways with the spectral change with high spatial- and temporal-resolution. This type of time-space-spectrum variable analysis will enable the visualization of molecular dynamics such as dissociation of proteins in biological reactions.
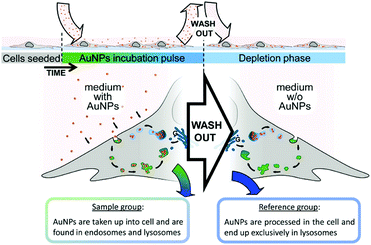 | ||
| Fig. 1 Schematic to illustrate the experimental design for characterization and visualization of vesicles in the endo-lysosomal pathway with SERS. (Adapted with permission from ref. 7. Copyright 2015 American Chemical Society.) | ||
2.2 Investigation of biological processes and cellular functions
Recent progress by applying SERS in investigating biological processes and cellular functions including cell secretion, cellular apoptosis, cell division processes, cellular stress, as well as cellular differentiation processes will be introduced. A nanosensor, which was designed by decorating AuNPs on borosilicate nanopipettes, was used to locally probe cellular secretion events near Madin-Darby canine kidney (MDCKII) epithelial cells as shown in Fig. 2.9 The selective detection of pyruvate, lactate, ATP, and urea was first demonstrated with low interference in a complex and biologically relevant medium based on a data-processing method capable of sorting and counting metabolites with reference to a SERS spectral database. In the next step, an increase in detection events for pyruvate was observed after the injection of saponin into the medium, showing that cells released a significant amount of pyruvate. This nanosensor can be a promising sensing tool for monitoring cell secretion events with temporal and spatial resolution.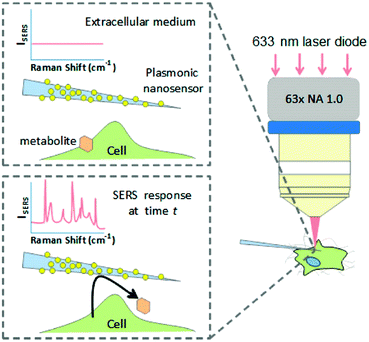 | ||
| Fig. 2 Schematic of the concept of an Au nanoparticle decorated nanopipette to monitor cellular secretion. (Adapted with permission from ref. 9. Copyright 2016 American Chemical Society.) | ||
A further study focused on the investigation of various biological processes and cellular functions at a molecular level by using functionalized nuclear-targeted nanoparticles for SERS experiments. First, the authors used nuclear-targeted AuNPs to monitor the morphological and molecular events during cellular apoptosis in real-time after the treatment of an apoptosis-inducing agent (see Fig. 3(a)).10 Real-time monitoring of the structural changes at a molecular level was achieved by taking the plasmonically enhanced Rayleigh scattering images and the SERS spectra dominated by the contributions of molecules around the cell's nuclear region targeted with functionalized AuNPs into consideration. The results are depicted in Fig. 3(b). Spectroscopic analysis of the occurrence and dynamics of three apoptotic molecular events, protein denaturation, proteolysis, and DNA fragmentation, were used to create a temporal profile of apoptotic molecular events in single cells. Moreover, nuclear-targeted gold nanocubes (AuNCs) are used in combination with SERS to probe the complex and dynamic biological processes of various stages of mitosis in cancer and healthy cells from a molecular perspective.11 The results show that a relatively high rate of conversion of mitotic proteins from their α-helix structure to β-sheet conformation is likely in cancer cells during the meta-, ana-, and telophases. Unique biochemical modifications of the lipid and amino acid moieties, associated with the observed protein conformational modifications, were also identified. However, in healthy cells, the existence of proteins in their β-conformation was momentary; mainly the α-helix form is present. This study could provide new insights into the role of protein conformation dynamics during mitosis in the development of cancer and many other diseases. In further work,12 SERS-based nuclear-targeted AuCNs were also used to elucidate the cellular responses of cancer cells towards various ultraviolet (UV) irradiations in real-time. Recently, the authors also used nuclear-targeted AuNPs to induce plasmonic photothermal therapy (PTT) cell death upon laser exposure, and simultaneously monitored the associated molecular changes in real-time using SERS within a single cell.13 The heat generated from the aggregated nanoparticles absorbing near-infrared laser light when irradiated with a sufficient laser power was found to cause modifications in the protein and lipid structures within the cell and, ultimately, this led to cell death. This study may help understand the molecular mechanism of PTT induced cell death, which is crucial to understand the mechanism of treatment and optimize its efficacy.
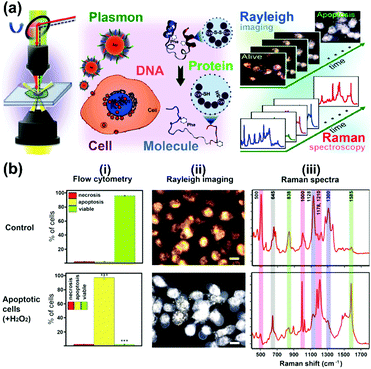 | ||
| Fig. 3 (a) Schematic of observing real-time molecular event dynamics of apoptosis in living cancer cells using nuclear-targeted plasmonically enhanced Raman nanoprobes. (b) Viable and apoptotic cells produce different images and spectra: (i) flow cytometric analysis; (ii) plasmonically enhanced Rayleigh scattering images; (iii) plasmonically enhanced Raman spectra of untreated viable cells (Control) and apoptotic cells treated with 100 μM H2O2. (Adapted with permission from ref. 10. Copyright 2014 American Chemical Society.) | ||
Rat pheochromocytoma PC12 cells are frequently used as a dopaminergic neuron model due to their various functions, including the synthesis, storage, and secretion of catecholamines. Furthermore, PC12 cells release a measurable amount of dopamine (DA) in response to some chemicals. Therefore, PC12 cells are considered to be one of the most common in vitro models for studying neurotransmitter release. SERS was applied to determine the in situ short-time effects of cisplatin, bisphenol-A, and cyclophosphamide on the extracellular DA level released from PC12 cells with high sensitivity by using an Au nanorod coated ITO substrate.14 The results demonstrated that the changes in the SERS spectra depending on the treatment agent were in agreement with the HPLC results on the extracellular DA level. In further work,15 the in vitro stepwise differentiation process of isolated mouse neural stem cells, the one-step differentiation of adult human neural stem cells (HB1.F3) and the electrochemical stimulation of PC12 cells were investigated by using a cell-based chip. The results showed that each cell line exhibited a different behaviour during differentiation. The DNA contents changed irregularly during the differentiation of HB1.F3 cells, while the percentage of proteins increased. In addition, the results revealed that the electrochemical stimulation of PC12 cells induced changes in the synthesis of DNA and proteins. The differentiation of isolated mouse neural stem cells showed a decrease in some peaks corresponding to the DNA content and an increase in the percentage of protein. These results demonstrate that the SERS technique allows for rapid biological analysis to monitor engineered tissues and optimize culture conditions in a real-time manner.
In conclusion, SERS has been a powerful tool for real-time and in situ investigations of various biological processes and cellular functions such as cell secretion, cellular apoptosis, cell division processes, cellular stress, and cellular differentiation. The obtained dynamic molecular information at a single-cell level may contribute to a better understanding of the nature of cell functions and enable a detailed and complete understanding of the pathogenesis of diseases.
2.3 SERS imaging of cell-surface species
The type and expression of cell-surface species are usually indexes of various diseases especially in the case of cancer. Imaging of cell-surface species with high specificity and sensitivity is essential for studying cellular processes and provides additional information for accurate diagnosis. SERS-based cell imaging offers excellent resolution and visualization for monitoring or tracking the cell-surface species. More importantly, the hyperspectral data received by imaging enable a reliable analysis, which is more significant than investigating only single spectrum in cellular studies.A biorthogonal SERS imaging approach that exploits small Raman reporter molecules was developed for visualizing cell-surface biomolecules.16 Biorthogonal Raman reporter molecules are small in size having azides, alkynes, and carbon–deuterium bonds, which are not present in biological systems. Cells that are metabolically labelled with these reporter molecules were cultured on a nanoparticle coated substrate and SERS mapping was performed. Various cell-surface biomolecules including proteins, glycans, and lipids were metabolically incorporated with the corresponding precursors bearing a Raman reporter molecule and visualized by SERS microscopy. The coupling of SERS microscopy with bioorthogonal Raman reporters expands the capabilities of live-cell microscopy beyond the modalities of fluorescence and label-free imaging. Glycosylation is one of the most common post-transcriptional modifications of proteins in eukaryotes. In situ visualization of glycans on specific proteins may provide the correlation of protein glycosylation with disease states. A zone-controllable SERS approach was developed for Raman imaging of protein-specific glycosylation on a cell surface using two types of newly designed nanoprobes.17 The signal probe, prepared using a Raman reporter molecule and dibenzocyclooctyne-amine to functionalize a 10 nm Au nanoparticle, exhibits a negligible SERS effect and can recognize and link the azide-tagged glycan via a click reaction. The substrate probe, aptamer modified 30 or 40 nm AuNPs, can specifically recognize the target protein to create an efficient SERS zone on the target protein. By controlling the size of the substrate probe to match the expression zone of the protein-specific glycan, an efficient SERS signal can be generated as shown in Fig. 4. This method has been successfully used for in situ imaging of sialic acids on the target protein epithelial cell adhesion molecule (EpCAM), a surface protein of MCF-7 cells, as well as for the monitoring of the expression variation of protein-specific glycosylation during drug treatment. This protocol is promising in uncovering glycosylation-related biological processes. Recently, the same research group showed that benzoic group functionalized gold nanoflowers can be used as a bridge probe for both recognition of the target sialic acid and assembly of poly(N-acetylneuraminic acid) modified AuNPs.18 Sensitive SERS imaging of sialic acids on the surface of living cells can be realized via the plasmonic coupling of these two kinds of nanoprobes in a single core-multi satellite nanostructure.
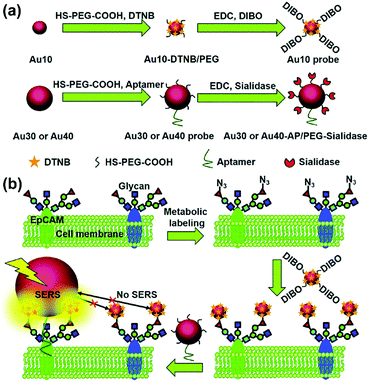 | ||
| Fig. 4 Schematic of (a) the structures of two types of Au nanoprobes and (b) the zone-controllable SERS effect for imaging of protein-specific glycans on the cell surface. (Adapted with permission from ref. 17. Copyright 2016 The Royal Society of Chemistry.) | ||
In conclusion, the visualization of expression and distribution of different types of cell-surface species including proteins, glycans, lipids, and sialic acids, has been realized by using SERS imaging based on the smart design of SERS tags with high sensitivity and specificity. Moreover, the expression variation of protein-specific glycosylation on a cell surface during drug treatment can be monitored. These protocols can be promising for the investigation of cell-surface species related biological processes and diseases.
3 SERS-based sensing of the cellular environment
In biological systems such as cells, subtle changes in the biochemical environment, such as pH, gaseous content, the content of reactive oxygen species (ROS) and redox potential, can have dramatic effects on cellular functions, and can cause diseases. Therefore, sensitive and reliable detection of the cellular microenvironment can facilitate the understanding of the changes during the occurrence and development of diseases and corresponding pathogenesis at a single-cell level. As a highly sensitive optical technique, SERS has received more and more attention in sensing the microenvironment of living cells. In this section, recent progress in SERS-based cellular microenvironment sensing will be highlighted regarding intracellular pH sensing, cellular gaseous sensing, as well as cellular ROS and redox potential sensing.3.1 Cellular pH sensing
Local pH of the cellular compartments is one of the most important regulating factors in the physiological activities of cells. SERS-based pH sensing is generally realized with a SERS-based pH nanosensor, in which the probe molecules, exhibiting both sensitive pH response and strong SERS signals, bind to the nanoparticle surfaces. With exposure to the environment with different pH values, the probe molecules experience protonation and deprotonation, resulting in obvious changes in the relative intensity of the SERS spectra reflecting the structural change. When internalized into the cells, the pH sensor can produce different SERS signals in response to the cellular microenvironment for different pH values. In this way, the local pH changes in the microenvironment can be monitored. Conventional nanoparticle-based SERS tags for pH sensing often fail due to the aggregation of particles when detecting in acidic medium or samples with high ionic strength. Therefore, many efforts have been made to improve the stability and reliability of SERS-based pH sensors.A SERS-pH sensitive substrate by using a novel Raman reporter, arene chromium tricarbonyl linked aminothiophenol (Cr(CO)3-ATP), functionalized onto a gold coated planar substrate with nanoscale roughness was developed for stable biological pH sensing.19 This new Raman reporter enables pH sensing by monitoring the shift of the Raman marker modes in a spectral range instead of peak intensity, which is relatively free of the peaks from biomolecules. The usage of a planar substrate helps in overcoming the limitation of the NP aggregation. This pH sensitive substrate exhibited high stability, and better sensitivity in the acidic pH range and has been applied for the pH sensing of a clinical urine sample with great accuracy as a proof-of-concept. This stable pH sensitive substrate holds the potential for extracellular pH sensing as well as pH sensing of bio-fluids for an early diagnosis. In addition to the planar pH sensitive substrate, p-aminothiophenol (pATP) functionalized Au@Ag/multi-walled carbon nanotubes (MWCNTs) were also designed for SERS-based pH sensing of living cells with a wide dynamic pH range.20 By using the MWCNTs as the substrate the state of aggregation of Au@Ag NPs is controlled, and the pH response range is extended from pH 3.0 to pH 14.0, which is much wider than the one using unaggregated Au@Ag NPs without MWCNTs. The reason is that the extent of aggregation of the Au@AgNPs can affect the transformation efficiency of pATP between the two states of protonation and deprotonation. Moreover, the MWCNTs can prevent further aggregation of Au@Ag NPs in the physiological environment, which ensures the stability and can be used for the study of physiological processes.
Different from the above mentioned solid substrates, the most widely used pH nanosensor is usually based on colloidal nanoparticles. However, there are still challenges concerning the reliable pH sensing of living cells by using nanoparticle-based pH sensors. To address these challenges, a facile way to prepare a robust pH nanosensor by coating the surface of the nanosensor with a protective molecule layer (BSA) was developed for a reliable intracellular pH analysis as illustrated in Fig. 5.21 AuNPs were used as the SERS-active core, while as the reporter molecule, 4-MPy was functionalized on the surface of the AuNPs. The protein BSA was subsequently immobilized on the surface of the pH-sensitive nanosensor to form a protective molecule layer. The BSA coating not only provides the pH nanosensor with a highly sensitive response to pH changes ranging from pH 4.0 to 9.0 but also exhibits high sensitivity, stability, reliability, and good biocompatibility in various solutions of high ionic strength or with complex composition, both in the aggregation state and after long-term storage. This pH nanosensor shows great advantages for a reliable intracellular pH analysis and has been successfully used to monitor the pH distribution of living cells providing high quality images of an intact cancer cell.
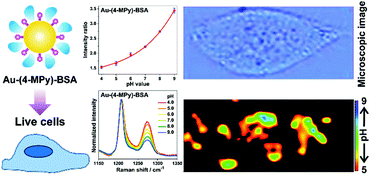 | ||
| Fig. 5 A BSA-coated pH nanosensor for improved SERS-based intracellular pH sensing. (Adapted with permission from ref. 21. Copyright 2014 American Chemical Society.) | ||
3.2 Cellular gaseous sensing
Carbon monoxide (CO) and nitric oxide (NO) are significant gaseous signalling molecules in various pathological and physiological pathways and have also drawn attention as promising therapeutic reagents for various human diseases. However, many aspects of their functions are still indistinct due to the shortage of powerful detection approaches. Similar to SERS-based pH sensing, SERS-based CO or NO sensing can be realized by using reporter molecules that can react with CO or NO to functionalize the SERS tags. Therefore, SERS-based nanosensors have recently found important applications in cellular gaseous sensing.A novel SERS-based nanosensor for the detection of CO based on the reaction of palladacycle carbonylation was designed.22 The newly synthesized palladacycles (PC) were assembled on the surface of AuNPs as the reporter molecule with good SERS activity and reactivity with CO. When the AuNP-PC nanosensors were incubated with a CO-containing sample, carbonylation of the PC was initiated, and the corresponding SERS spectra changed significantly, which allowed the carbonylation reaction to be directly observed in situ as shown in Fig. 6. The nanosensor also exhibited high selectivity over other biologically relevant species. Benefiting from the specific features of the PC carbonylation and the superior spectral resolution of SERS, the detection of CO in living cells was realized without interferences from other biological species.
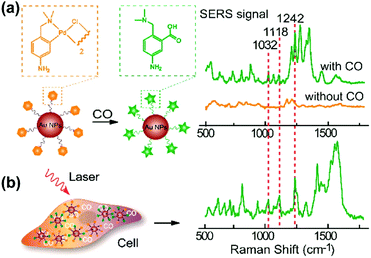 | ||
| Fig. 6 (a) SERS response and sensing mechanism of palladacycle carbonylation on AuNP/PC nanosensors for CO. (b) SERS detection of CO in living cells. (Adapted with permission from ref. 22. Copyright 2015 American Chemical Society.) | ||
Regarding NO sensing, o-phenylenediamine (OPD)-modified AuNPs were designed as a new kind of SERS tag for detecting endogenous NO in living cells rapidly, sensitively and selectively.23 Through the OPD modification onto the surface of AuNPs, the NO reactivity is successfully combined with a strong SERS activity. When incubated with NO-containing samples, the diamine moieties of OPD molecules may turn into triazole moieties in the presence of O2 on the surface of AuNPs, which can be sensitively detected by the corresponding change in the SERS spectra. By using this SERS-based NO sensor, NO in solution can be detected rapidly within 30 s with a sensitivity of 10−7 M. Notably, the real-time and in situ monitoring of NO produced by the iNOS pathway in a living macrophage stimulated with lipopolysaccharide (LPS) was also demonstrated by using these nanosensors.
3.3 Cellular reactive oxygen species and redox potential sensing
The ROS content is an important physiological index of the cellular microenvironment, which refers to the oxygenic radicals derived from the oxygen metabolism including ˙OH, H2O2, ROO˙, singlet oxygen, hypochlorous acid etc. ROS are involved in various physiological activities such as cell signalling, gene expression regulation and immune reactions. However, overproduction of ROS can also cause oxidative damage to the cell structures including lipid and protein membranes, proteins and DNA, and therefore lead to various diseases. SERS-based nanosensors can also be applied to monitor cellular ROS.Detection of H2O2 with high sensitivity and selectivity in living cells is a challenge for evaluating the diverse roles of H2O2 in the physiological and pathological processes. To address this, a SERS nanosensor was developed for the sensitive and selective sensing of H2O2 in living cells by functionalizing 4-carboxyphenylboronic acid (4-CA) on the surface of AuNPs (AuNPs/4-CA).24 This nanosensor was designed based on the specific transformation of arylboronate into phenol in the presence of H2O2, which leads to SERS spectral changes of AuNPs/4-CA. In Fig. 7(a), a schematic description of the sensor platform and the corresponding SERS spectra are given. This SERS-based nanosensor exhibits high sensitivity and selectivity for H2O2 over other reactive oxygen species, abundant competing cellular thiols as well as biologically relevant species. In addition, the AuNPs/4-CA nanosensor shows long-term stability against incubation time and pH, and high biocompatibility, which has been successfully applied to detect the level of H2O2 in living cells under oxidative stress. Furthermore, a boronate nanoprobe based on 3-mercaptophenylboronic acid (3-MPBA) functionalized AuNPs was also developed for quantitative, selective, and sensitive detection of H2O2 over other ROS in PBS with a LOD of 70 nM, which has been applied to detect exogenous and endogenous H2O2 in living cells.25
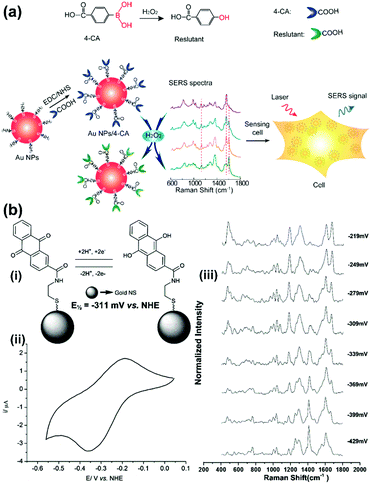 | ||
| Fig. 7 (a) Illustration of the AuNPs/4-CA nanosensors for sensing H2O2 in living cells (Adapted with permission from ref. 24. Copyright 2015 Elsevier B.V.) (b) SERS nanosensors for quantitative measurement of redox potential: (i) structures, electron transfer schemes and standard reduction potentials; (ii) cyclic voltammogram of reporter molecule; (iii) potential-dependent changes in SERS spectra. (Adapted with permission from ref. 27. Copyright 2014 The Royal Society of Chemistry.) | ||
Cellular redox potential refers to the oxidative or reductive state in a living cell, which plays an important role in controlling pathway activation via modulation of the oxidation state of proteins. Redox potential changes are associated with various cellular processes such as cell cycle, differentiation, apoptosis and signalling, and have demonstrated that the dysfunction of the redox potential in living cells can play a decisive role in the initiation or progression of diseases including cardiovascular disorders, neurodegenerative diseases and cancer.
A single cell-based chip was designed to investigate the intracellular and extracellular redox state of PC12 cells using SERS in combination with linear sweep voltammetry (LSV).26 PC12 cells were immobilized on gold nanodots/ITO surface and subjected to LSV while the intracellular biochemical changes were successfully monitored by SERS simultaneously. Moreover, paired gold microelectrodes with a micrometer-sized gap containing hexagonal arrays of gold nanodots were fabricated to detect electrochemical activity and changes in the redox environment of a single PC12 cell. Organic reporter molecules that showed oxidation-state-dependent changes in the Raman spectra were modified onto the surface gold nanoshell to form a SERS-based nanosensor for the quantitative measurement of the redox potential in a hypoxic cell (see Fig. 7(b)).27 By introducing the nanosensors into cells and collecting the SERS spectra, the corresponding localized intracellular redox potential from single hypoxic cells can be calculated in a non-invasive, reversible way. In further work,28 the authors demonstrated that the SERS-based intracellular nanosensors could be improved to cover a much wider redox potential range of −470 to +130 mV vs. NHE which includes the redox potential range occupied by cells in a state of oxidative stress. The nanosensors are rationally designed to target different sections of this redox potential range and are monitored by SERS which will permit real-time imaging of cells undergoing oxidative stress.
To summarize, by functionalizing SERS tags with different specific sensing reporter molecules, the SERS-based nanosensors are successfully applied with high sensitivity for sensing the changes in the cellular microenvironmental parameters, such as pH, gaseous content, as well as the content of ROS and redox potential. More emphasis is given on improving the sensitivity, selectivity, and stability to ensure reliable sensing in living cell systems taking into account the complexity of the cellular microenvironment. The reliable SERS-based sensing strategies can be applied to monitor the changes within the microenvironment in various biological processes, which may help in understanding the pathogenesis of diseases at a single-cell level.
4 SERS as a diagnostic tool for tumour identification and in vivo cancer detection
The clinical potential of SERS is evident as already introduced; however, studies to identify tumour margins or to detect cancer in vivo have not been discussed in this review article so far. Therefore, this section summarizes recently published work for the differentiation and identification of cancer cells, the application of SERS toward the detection of CTCs, the investigation of tissue sections to identify tumours for surgical guidance and, finally, the in vivo detection of cancer is discussed.4.1 SERS-based detection and identification of cancer and circulating tumour cells
The high prevalence of cancer and the increased survival rate after early diagnosis motivate the development of specific and sensitive methods such as SERS as powerful tools for early cancer diagnosis. As one example, cancer cells are exfoliated from tissue and transferred onto a SERS active substrate.29 Reproducible SERS spectra are recorded for normal and cancerous cells and the discrimination is achieved by analyzing band ratios. For the investigation of 20 patients a sensitivity and specificity of 100% has been demonstrated, respectively.Since the label-free SERS spectra are very complex and a full band assignment is often not possible, SERS tags were applied for indirect detection of cancer cells. Thus, the spectral response is clearly assignable to the targeted cancer cells based on the marker modes of the reporter molecule. In order to allow for a rapid identification with high throughput, a microfluidic channel system was applied for the detection of cancer cells in low concentrations targeted by SERS labels.30 By applying SERS tags a unique SERS spectrum was recorded in the presence of the cancer cell, which can be deconvoluted into a complex spectral response. The identification of individual targeted cancer cells was correctly performed in the background of a large number of other cells. The authors demonstrated that the application of two different SERS tags indicating the overexpression of neuropilin-1 and the unspecific binding toward the cell membrane, respectively, combined with a deconvolution strategy results in a detection limit of 1 cancer cell in 100 other cells.
The concepts employing SERS tags, which are developed employing cultured cancer cell lines, were further modified to enable the detection of CTCs. The presence of CTCs is related to the occurrence of a primary tumour and/or metastasis. Since their concentration in blood is very low, the application of signal enhancing techniques is needed in order to detect them among millions of leukocytes and billions of erythrocytes. To allow for the best possible signal intensity, differently shaped gold nanoparticles were modified with a reporter molecule as well as folic acid.31 Thus, specific interaction with tumour cells was achieved due to the overexpressed folate receptor alpha. Under optimized measuring conditions, CTCs were detected in blood without further enrichment and the achieved LOD is 1 cell per mL. Moreover, the detection of single breast cancer cells in unprocessed blood has been demonstrated by using an antibody rainbow cocktail, i.e. silver–gold nanorod hybrid structures modified with four different reporter molecules and four antibodies specific for breast cancer as well as with a leukocyte-specific CD45 marker.32 The authors concluded that the detection of a single cancer cell within 7 million blood cells shows the high potential of the proposed protocol for an accurate and early detection of cancer to enable the subsequent well-timed treatment. In summary, the detection of single cancer cells is of great importance for the early detection of cancer and a successful assessment is necessary for the treatment of cancer. As a further strategy, sample purification steps e.g. by using magnetic nanoparticles in order to separate the captured cells from the background will gain importance.
4.2 Identifying tumour margins by means of SERS
Once the tumour is developed and surgery is necessary to save the patient's life, a surgical guidance becomes important in order to assess the completeness of the removal of the cancerous tissue. Since the expression of biomarkers is heterogeneous, molecular imaging of different cancer biomarkers in a multiplexed manner should be addressed. Therefore, SERS-based detection schemes are available and will be discussed in the following paragraph.After the surgical removal of human thyroid tissue, the samples were sliced and silver nanoparticles (AgNPs) were directly coated onto the tissue surface.33 To discriminate between different tissue types, i.e. thyroid cancer, nodular goiters as well as normal thyroid tissue, the recorded SERS spectra were analysed by PCA in combination with LDA. The authors achieved, for the discrimination of the three types of tissue, sensitivity values between 75 and 92% and for specificity, values between 82.6 and 89.4% were found. Thus, label-free SERS can be applied for the assessment of the tissue type at a molecular level. Moreover, a silver nanostructured surface is generated and tissue sections from oral cancer patients are prepared on them.34 The study contains 24 normal and 32 tumour tissue sections acquired from 8 oral cancer patients. Again, the SERS spectra are analysed by PCA and discriminant analysis resulting in a sensitivity of 100% and a specificity of 95.8%.
To identify residual tumours and their margins and to support the surgeon with information enabling the complete removal, SERS tags with different cancer related antibodies are applied addressing the heterogeneous expression of cancer biomarkers such as the epidermal growth factor receptor (EGFR) and human epidermal growth factor receptor 2 (HER2).35 To allow for quantitative molecular phenotype (QMP) mappings, a SERS tag modified with an isotype control antibody was applied in addition, and the band ratio of both marker modes referring to the cancer biomarker and to the control was plotted. Employing normal tissue samples and tumour xenografts with different EGFR expression levels the positive binding events of the EGFR SERS tags are illustrated. The clinical potential of the proposed method was shown by investigations of human breast tissue stained with HER2 and isotype SERS tags. The results are presented in Fig. 8. The nanoparticle concentration for both normal and tumour tissue is estimated and due to the specific interaction of the HER2 SERS tags with the tumour tissue, the QMP ratio allows for distinguishing between both sample types. The QMP images illustrate the identification of junctions of HER2-postive tumour and normal tissue. The same approach employing targeted EGFR and untargeted isotype SERS tags was applied for the detection of human glioma and epidermoid tissue.36 The binding potential of EGFR nanoparticles was estimated for healthy and tumour sections and based on these values, the successful identification of tumour margins was evident.
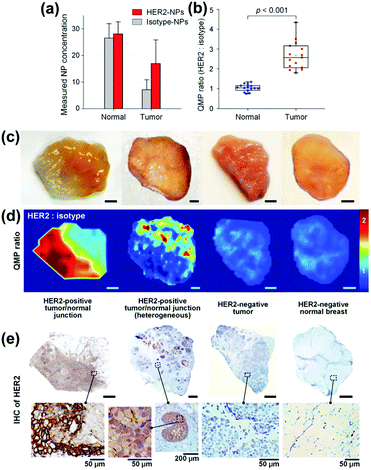 | ||
| Fig. 8 Quantitative molecular phenotype (QMP) imaging of breast cancer tissue. (a) Estimated concentrations of the SERS tags as well as (b) band intensity ratio for both tumour and normal tissue. (c) Photographical images of patient samples. (d) QMP ratio plots illustrating HER2-positive tumour areas. (e) Immunohistochemistry staining as control experiment. (Adapted with permission from ref. 35. Copyright 2016 Macmillan Publishers Limited.) | ||
In conclusion, label-free SERS is successfully applied to discriminate between tumour and healthy tissue. Due to the complexity of the recorded SERS spectra PCA and discriminant analysis are applied in order to achieve reliable results. Moreover, the application of SERS tags labelled for the specific detection of cancer biomarkers such as EGFR and HER2 in comparison to untargeted isotype SERS tags illustrates the clinical potential for surgical guidance in cancer therapy. Tumour margins as well as heterogeneous junctions of tumour and normal tissue were correctly identified.
4.3 In vivo detection of cancer
Based on the scientific achievements in the detection of cancer cells and tumour tissue, the next consequent step in bringing SERS towards applications was to study the in vivo detection of cancer. In doing so, SERS tags showing excellent signal intensity are applied equipped with specific recognition elements allowing for a specific interaction of overexpressed cancer biomarkers. The Raman reporter molecules should show a unique fingerprint spectrum, which can be easily separated from the spectral background due to the complex biological matrix.Due to their excellent plasmonic behaviour allowing for strong SERS signals of the Raman reporters, gold nanostars were used to illustrate the potential of SERS for in vivo investigations.37 Employing NIR excitation, the penetration of light through the skin and therefore, the SERS-based detection of the nanoparticles modified with a Raman reporter were achieved for 300 and 600 μm thick rat skin grafts. The authors demonstrated the ex vivo SERS signal detection of dye-labelled gold nanostars injected intradermally into rat dorsal skin after explantation. Finally, scaffolds loaded with dye-labelled gold nanostars were implanted into pig dorsum and SERS spectra were recorded after 1, 2, 3 and 6 days illustrating the potential for long-term sensing applications. In a further study, gold nanostars were applied as contrast agents and the functionalized nanoparticles were injected into mice with a SKOV3 xenograft tumour.38 To validate the size of the tumour, magnetic resonance imaging and bioluminescence imaging were performed. The nanoparticles accumulated passively in the tumour and the authors found no acute effect on the animal's health. SERS spectra were recorded three hours after the injection and the marker mode of the Raman reporter molecule was clearly identified. The results were confirmed by ex vivo SERS imaging and dark field microscopy after sacrificing the mice and the preparation of tissue slices.
SERS tags were applied being equipped with Raman reporters, targeting the antibody (anti-EGFR) as well as NIR fluorophores allowing for flow cytometry.39 The authors illustrated in in vitro experiments the capability of the proposed detection platform for specific targeting of lung cancer cells as well as the correlation of the SERS results with fluorescence intensities measured applying flow cytometry. For in vivo experiments, a cocktail containing three different SERS tags, i.e. two SERS tags for the specific targeting of EGFR but differently labelled with Raman reporters and a control SERS tag without a specific recognition element, was applied on a xenograft tumour after careful removal of the overlying skin. The incubation was performed for 30 min and after several washing steps the SERS images were recorded. For both EGFR-targeting SERS labels, the same signal distribution was achieved and no unspecific interaction of the control SERS tag with the tissue was found as illustrated in Fig. 9. The signal intensity pattern being heterogeneous across the tumour surface was confirmed by ex vivo staining using conventional immunohistochemistry.
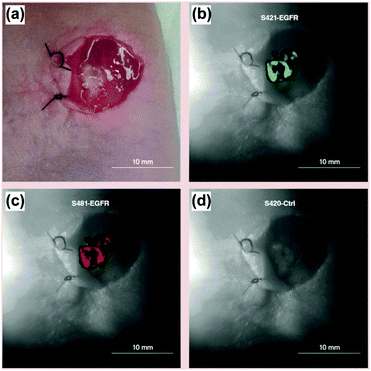 | ||
| Fig. 9 In vivo and multiplexed targeting of tumour tissue. (a) Colour image of a xenograft tumour after removal of the skin. White-light image of the tumour superimposed with the distribution of the marker mode of (b and c) two EGFR targeting SERS tags as well as (d) the unspecific control probe. (Adapted with permission from ref. 39. Copyright 2015 Future Medicine Ltd.) | ||
Endoscopic imaging or data acquisition allows for minimally invasive investigations in real-time clinical applications. To enable a specific and multiplexed detection of pathologic lesions, a fluorescence-SERS tag was developed.40 Employing an optical fibre bundle for illumination and detection of fluorescent as well as Raman scattered light, both EGFR and HER2 targeting nanoparticle arrangements, based on the unique fingerprint of the reporter molecules, were detected within mouse bearing cancer xenografts with different levels of free EGFR and HER2 biomarkers overexpressed in breast cancer cells. Moreover, the design, development and in vivo demonstration of an endoscopic system were shown for an intraluminal, multiplexed imaging of differently labelled SERS tags.41 Due to the application of different reporter molecules the multiplexed detection and the ratiometric analysis of the SERS spectra for a quantitative assessment are feasible. The in vivo demonstration of the endoscopic system was performed using an oesophageal mucosal layer of a post-mortem pig after the injection of 3 differently labelled SERS tags. This endoscopic SERS approach was further developed towards the detection of oesophageal cancer by means of topically applied SERS tags.42 As illustrated in Fig. 10, a tumour model was prepared by surgical attachment of a tumour into a rat's oesophagus. In the next step, SERS tags with specific antibodies to enable selective binding to the tumour site were applied, and localization and identification were achieved by endoscopic imaging within the oesophagus. By means of using additional isotype SERS tags, the unspecific interaction with the tissue is levelled out and the SERS-based identification of the tumour is achieved by a ratiometric approach, i.e. calculating the SERS ratio of the biomarker targeting SERS tag, binding to overexpressed EGFR or HER2, related to the SERS intensity gained from the isotype SERS tag. As this proposed detection scheme is also shown on tumour sections,35,36 its application for the in vivo detection of cancer due to topically employed SERS tags is the next consequent step toward a real clinical application. The authors concluded that SERS tags with improved optical characteristics are needed to achieve a better signal to noise ratio and to allow for shorter integration times.
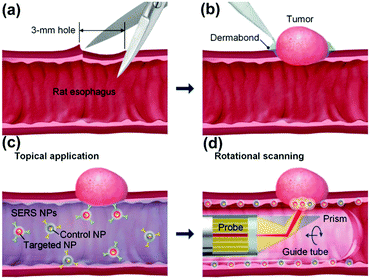 | ||
| Fig. 10 Schematic description of the endoscopic detection of a tumour model in a rat's oesophagus. (a and b) Surgical attachment of the tumour to the oesophagus. (c) Application of SERS tags on the luminal surface. (d) Endoscopic detection of the SERS signal originating from the SERS tags. (Adapted with permission from ref. 42. Copyright 2015 OSA.) | ||
To summarize, in vivo SERS-based procedures are available for the detection of tumour models in small animals or after removal of the skin as well as employing endoscopic approaches. All of the discussed papers applied a common 785 nm laser line for the SERS measurements. This is due to the tissue optical window allowing for penetration depths of several 100 μm into tissue. Thus, the SERS method can be used for tumour detection on the surface or underneath the skin; however, tumours buried in tissue cannot be monitored in 3D by means of the application of SERS tags. Therefore, SERS needs to be combined with imaging techniques such as magnetic resonance imaging if 3D monitoring of cancer within the human body is aimed for.
5 Theranostics
The term theranostics is created from therapy and diagnostics. Theranostics defines the increasingly closer link between diagnostics and therapy and aims for the correct therapeutic measure for the individual patients at the desired time. The development of nanomaterials with unique optical, plasmonic, electronic, magnetic, and multifunctional properties has opened up new possibilities for spectroscopic detection and molecular imaging in theranostics applications. Recently, SERS guided theranostic nanoplatforms based on smart designs of SERS tags for multifunctionality and in combination with other imaging techniques for multimodal approaches have attracted considerable attention. A drug-loaded nanomaterial called nanomedicine has attracted increasing attention since it can provide personalized theranostics with improved treatment efficacy and specificity. Multimodal imaging providing complementary information can dramatically improve the accuracy of diagnostics, which has tremendous potential in pre-clinical research and translational imaging. Furthermore, multifunctional theranostic platforms that generally integrate targeting, drug delivery, imaging and therapy into one nanoplatform are of great importance for high-performance theranostics. In this section, the recent progress in the monitoring of drug release, multimodal imaging theranostic strategies, and multifunctional theranostic platforms will be reviewed.In situ monitoring of drug release in cancer cells is essential for real-time assessment of drug release dynamics in chemotherapy. To do so, label-free in situ monitoring and control of the intracellular anti-drug delivery process was proposed by using SERS-based biohybrid nanoparticles.43 The biohybrid nanoparticles consist of a AuNP for SERS activity, doxorubicin (DOX) as the anti-cancer drug, a cell-penetrating peptide for increased uptake, and a cancer-targeting antibody for specific targeting, respectively, as shown in Fig. 11(a). The releasing rate of DOX was controlled with the addition of glutathione (GSH) on the cells and the intracellular release of DOX from the biohybrid nanoparticle was continuously monitored with time-dependent change of intracellular SERS signals of DOX (see Fig. 11(b–e)). The HER2-positive cancer cell was specifically targeted with the biohybrid nanoparticles and 90.23% of DOX was released by the action of intracellular GSH after 24 h treatment. The proposed strategy can also be applied for label-free, in situ monitoring of the time-dependent release of other anti-cancer drugs in living cells.
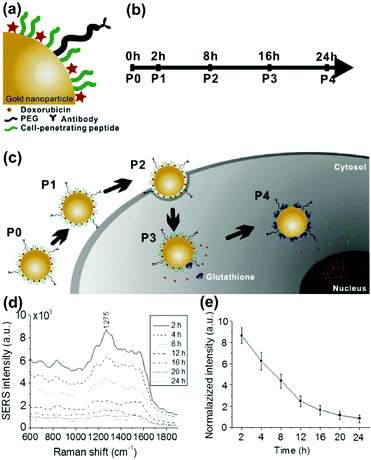 | ||
| Fig. 11 Schematic diagram for (a) conjugation of the biohybrid nanoparticle, (b) time-dependent monitoring of the nanoparticle's specific targeting, cellular uptake, and drug release, and (c) uptake of the DOX-loaded biohybrid nanoparticles by the cells and the intracellular release of DOX by GSH. (d) SERS spectra of the SK-BR-3 cells treated with DOX loaded biohybrid nanoparticles at different incubation times. (e) Relationship between time and release of DOX. (Adapted with permission from ref. 43. Copyright 2015 Elsevier B.V.) | ||
In addition to monitoring the drug release in cancer cells, monitoring the molecular changes of cancer cells with drug treatment is also crucial for the design of new anti-cancer drugs, the development of novel diagnostic strategies, and the improvement of the cancer therapy efficiency. A strategy that combined dark-field and fluorescence co-imaging assisted by means of SERS using nuclear targeting SERS tags was proposed for in situ monitoring of the molecular changes of drug-treated cancer cell nuclei.44 Two model drugs, a DNA binder (Hoechst33342) and an anti-cancer drug (DOX), were tested on the Soma Gastric Cancer cell (SGC-7901). In situ SERS spectra at the same positions with SERS tags in one cell nucleus were measured with the accurate location via dark-field and fluorescence imaging. By analysing the in situ recorded SERS spectra, it was found that both Hoechst33342 and DOX can cause obvious structural changes of DNA and proteins with different underlying mechanisms influencing the cell nuclei. Hoechst33342 tends to interact with DNA bases while DOX can induce DNA damage. These results may contribute to an improved understanding of the action effects of drugs.
A gold nanostar (GNS) probe was developed for multimodal imaging theranostics including SERS detection, CT, two-photon luminescence (TPL) imaging, and PTT.45 The biodistribution and intratumoral uptake of GNS probes were investigated at both macroscopic and microscopic scales by using optical imaging, radiolabeling, as well as CT. The ability of the GNS nanoprobes for in vitro photothermal heating and in vivo photothermal ablation of primary sarcomas in mice was also characterized. The GNS probe exhibited superior photothermal conversion efficiency due to a tip-enhanced plasmonic effect. In vivo PTT with a near-infrared (NIR) laser under the maximum permissible exposure (MPE) led to ablation of aggressive tumours containing GNS, but had no effect in the absence of GNS. This multifunctional GNS probe has the potential to be used for in vivo biosensing, preoperative CT imaging, intraoperative detection with optical methods (SERS and TPL), as well as image-guided photothermal therapy. Trimodal MRI-SERRS-fluorescence detection of cancer cells was realized by using magneto-plasmonic nanoprobes as shown in Fig. 12(a).46 The nanoprobes consist of SERS-active AgNPs, super-paramagnetic iron oxide nanoparticles for MRI capacity, nile blue as the fluorescent dye and a SERRS reporter molecule, as well as an encapsulation by the biopolymer chitosan for improved biocompatibility. Trimodal detection of HeLa cancer cells in tissue phantoms by using the magneto-plasmonic nanoprobes was demonstrated. A multimodal Raman and photoacoustic imaging system was designed to specifically trap and detect CTCs with modified SERS tags in vivo and to provide structural information of the surrounding vasculature.47 The results are depicted in Fig. 12(b). Photoacoustic imaging provided a microvascular context and can potentially be used to guide magnetic trapping of CTCs for SERS detection in animal models. This technique could be used for the investigation of CTCs, providing molecular information along with structural or functional vasculature imaging.
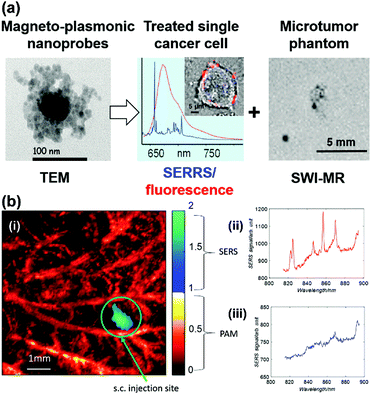 | ||
| Fig. 12 (a) Trimodal magnetic resonance imaging-SERS-fluorescence detection of cancer cells. (Adapted with permission from ref. 46. Copyright 2015 American Chemical Society.) (b) Multimodal Raman and photoacoustic imaging for in vivo studies: (i) rat ear vasculature imaging by photoacoustic imaging, and co-registered Raman imaging of injected targeted-HeLa cells; (ii) SERS spectrum obtained from the magnetic trapping region of a chicken embryo membrane and (iii) away from the magnetic trapping region. (Adapted with permission from ref. 47. Copyright 2016 SPIE.) | ||
From the perspective of clinical application, a real-time clinical endoscopic system was designed for intraluminal, multiplexed imaging of biomarker-targeting SERS tags in the human gastrointestinal tract for the early diagnosis of cancer.41 In Fig. 13, the detection scheme is shown. This system enabled rapid circumferential scanning of topologically complex luminal surfaces of hollow organs and produced quantitative images of the relative concentrations of SERS tags that are present. More importantly, it also offers unparalleled multiplexing capabilities by simultaneously detecting the unique spectral fingerprints of multiple SERS tags. This new screening strategy holds the potential to improve diagnosis and to guide therapy in the context of white-light endoscopy.
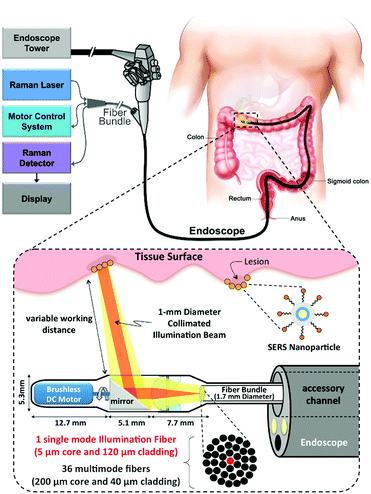 | ||
| Fig. 13 Schematic of Raman-imaging system being used in parallel with white-light endoscopy. (Adapted with permission from ref. 41. Copyright 2015 Garai et al.) | ||
Recently, a SERS-assisted theranostic strategy based on Fe3O4/Au cluster/shell nanocomposites was designed and used for PSA detection, MRI, and magnetic hyperthermia.48 SERS is sensitive for the diagnosis of the prostate cancer biomarker PSA with a LOD down to 0.75 ng mL−1 while MRI can be used to determine the location of a tumour. In addition to diagnosis, magnetic hyperthermia was also employed as a treatment and revealed a killing effect for the tumour cells. This SERS-assisted strategy might be integrated into diagnosis and treatment of tumours in future work. A yolk–shell multifunctional SERS tag, gold nanorod@void@mesoporous titania nanoparticle (AuNR@void@mTiO2 NP), was developed as a theranostic tool for simultaneous SERS imaging and chemophotothermal therapy (see Fig. 14(a)).49 This SERS tag consists of a NIR light absorbing AuNR core as the SERS-active substrate and photothermal agent and an mTiO2 outer shell for drug loading, which integrate multifunctionality into one SERS tag. The NIR light absorbing property of AuNRs was also explored for drug release regulation and photothermal treatment. Benefiting from the synergistic chemo-thermal therapeutic effect, a significant decrease in viability of MCF-7 cells was observed when treated together with DOX-loaded NPs and NIR laser irradiation. This multifunctional SERS tag might be a promising theranostic platform. A new type of 3D close-packed nanoassembly based on pluronic stabilized gold nanoaggregates of controlled size and labelled with methylene blue (MB) was designed for multimodal cell imaging and enhanced photodynamic therapy as illustrated in Fig. 14(b).50 The nanoassemblies enable the optical imaging of murine colon carcinoma cells (C-26) recording both Raman and fluorescence signals collected from MB molecules by using scanning confocal SERRS and fluorescence lifetime imaging microscopy (FLIM) techniques. Furthermore, the conjugation of MB molecules to the nanoassemblies can also improve the photodynamic therapeutic activity due to the enhanced singlet oxygen generation ability. This strategy based on combining SERS tags with reporter molecules, which exhibit both fluorescence and enhanced photodynamic properties, provides a new way to develop multifunctional theranostic platforms.
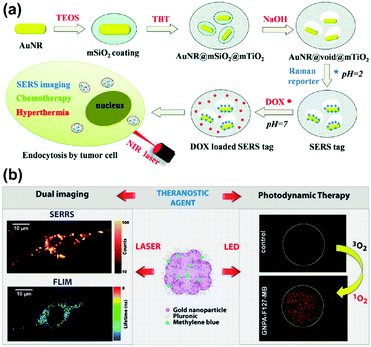 | ||
| Fig. 14 (a) Schematic of AuNR@void@mTiO2 yolk–shell NPs as multifunctional theranostic platforms for cancer treatment. (Adapted with permission from ref. 49. Copyright 2014 The Royal Society of Chemistry.) (b) Theranostic agents based on pluronic stabilized gold nanoaggregates loaded with methylene blue for multimodal cell imaging and enhanced photodynamic therapy. (Adapted with permission from ref. 50. Copyright 2015 American Chemical Society.) | ||
To summarize, SERS-based theranostic platforms have played an important role in the biomedical and clinical fields including the monitoring of nanomedicine-based drug release, as well as providing multimodal imaging and multifunctional capacity with improved accuracy of diagnosis subsequently with therapeutic possibility. More accurate diagnosis can be ensured based on the multi-parameter results compared with the conventional single parameter detection protocol. More importantly, SERS with multiplex detection and multimodal imaging abilities, integrated with in vivo endoscopic techniques, exhibits high potential to be a promising tool for guiding next generation surgical treatments.
6 Summary and perspective
In this review, recent progress in using SERS as a powerful analytical tool for biological and biomedical applications is summarized for selected topics on cells being the basic structure and functional units of organisms toward clinically relevant topics. The applications of SERS-based detection schemes in living cell studies are reviewed, including the direct intracellular analysis, investigation of biological processes and cellular functions, imaging of cell-surface species, as well as SERS-based sensing of the cellular environment (pH sensing, gaseous sensing, and redox condition sensing). In addition, from the viewpoint of practical and clinical applications for diagnosis and therapy, SERS-based diagnostic techniques including the detection and identification of cancer, CTCs, tumour margins, and in vivo detections are also reviewed. Finally, the theranostic capabilities of SERS for the monitoring of drug release, multimodal imaging theranostic strategy, and multifunctional theranostic platform are highlighted.For the SERS-based study on living cells, the detection of cancer cells or CTCs or for the identification of tumour margins, challenges are mainly faced and can be addressed in the following four aspects. First, the selective detection needs to be addressed, since a cell is much more complex than conventional pure samples, because all biomolecules present in cells can contribute theoretically to the SERS signals, depending on their affinity toward the metallic surface. In general, the usage of chemometrics for the data analysis is expected to be rising in the future due to the complexity of the label-free SERS spectra. Thus, trends toward the functionalization of the SERS-active substrate surface with appropriate functional recognition units to improve the selectivity are seen. Second, quantitative detection is of great importance in biology and medicine. However, in contrast to normal Raman, the proportionality between the intensity and the concentration of the analyte in SERS is not entirely linear due to e.g. non-reproducible SERS substrates or saturation effects. Fabrication of uniform, sensitive and clean SERS-active substrates or the introduction of innovative internal standards tends to be promising. Third, the spectral analysis should be addressed with more attention. Since the complex biochemical composition of cells results in SERS spectra with contributions of various biomolecules, the interpretation and extraction of useful information from the spectra are challenging. An alternative way is using multivariate algorithms, which can help simplify the spectral data and explain the spectra in a mathematical way, whereas reliable SERS spectra of some standard biomolecules such as amino acids and metabolites may be required as the reference for reliable assignments of some specific peaks. Finally, the reliability of the SERS results have to be assessed since a living cell is a dynamic system in time and space. So far, most of the studies on cellular detection have failed to obtain information of cells in their native state and reflect their real physiological activities. It's a promising trend to integrate in vivo cell culturing, microscopy imaging with multispectral measurements that employ innovative design of cell culturing chambers, and multifunctional nanoprobes for rapid, highly temporal and spatial detection. Moreover, microfluidic approaches will be gaining importance in future work in order to allow for high-throughput measurements with increased automation, which is enormously important for clinically relevant detection schemes e.g. for the detection of CTCs in blood samples.
In order to obtain a detailed understanding of the development of diseases and corresponding pathogenesis at the single cell level, monitoring of the biochemical environment will gain importance in future work. Here, SERS active nanostructures are applied which are equipped with specific molecules capturing small molecules or being sensitive to local changes of the pH value. Since the local pH value of cell compartments is one of the most regulating factors in physiological activities, nanosensors were developed and the attached probe molecule shows a different SERS response of the protonated and deprotonated form. Depending on the probe molecule, the pH sensing range can be varied. Due to decomposition processes in endosomes and lysosomes, the stability of those nanosensors can be increased by different coating materials, e.g. BSA or silica. It is expected that the measurement of the local pH value in cells will accelerate the fundamental research on cellular processes influencing the local pH value e.g. ion channel activity. The small molecules CO and NO are known as signalling molecules in various pathological and physiological pathways and have also drawn attention as a promising therapeutic reagent for various human diseases. Similar to the local pH value measurements, SERS nanosensors are designed with specific capture molecules and in the presence of the signalling molecules, the SERS spectrum is changed. Thus, successful detection is achieved even in a complex matrix such as cytoplasm. The same is performed for the intracellular detection of ROS species using reporter molecules showing oxidation-state-dependent changes in their Raman response. All intracellular detection schemes have in common the fact that insights into cellular activities can be drawn, which will accelerate the fundamental research to find new coherences between cellular processes at the single cell level and the development of diseases in future work. The great advantage of rationally designed nanosensors based on SERS is that the analyte of interest is captured specifically, and the SERS response is simplified since the molecular changes of the reporter molecule due to the interaction with the analyte dominate the SERS spectra. Unspecific interactions are suppressed and contributions from the complex matrix e.g. the cell interior are not or less contributing to the SERS signals. Thus, the spectral analysis will be mostly based on the change in the band ratio in order to quantify the analyte molecules and it is expected that complex chemometrical analyses are not necessary. Despite the merits of SERS-based sensing in living cells, it is of particular importance to ensure the reliability of the SERS results by considering the complexity of the cellular environment. This raises the demand and challenge on the rational design of future SERS-based nanosensors. In addition, as a matter of fact, all the microenvironmental parameters (pH, gaseous content, ROS and redox state etc.) in living cells coordinate and interact synergistically instead of individually. Any single parameter could be emphasized in a certain aspect but ignored in others. Therefore, to enable comprehensive and objective understanding of the intracellular microenvironment, a multifunctional nanosensor platform which can integrate the sensing of multiple parameters would be the future trend of SERS-based sensing in living cells. Moreover, tremendous opportunities are provided for the powerful design of SERS-based nanosensors with multiplexing capacity to monitor multiple cellular microenvironmental parameters.
The clinical potential of SERS is evident reflecting the results on the SERS-based investigations of living cells and the cell's intracellular environment. Consequently, the potential of SERS-based detection schemes was illustrated for the detection of CTCs, tumour margins for surgical guidance and the in vivo analysis of cancer. In the case of CTCs, the challenge is to find the needle in the haystack since only a few CTCs are found in 1 mL of blood. Therefore, powerful SERS tags with specific recognition units are of advantage to identify the CTCs in a background of blood or cell material. To allow for high-throughput measurements, the combination with microfluidic devices will increase in future work. Moreover, intelligent sample preparation protocols are necessary to bring SERS-based detection of CTCs into clinical application, e.g. by using magnetic nanoparticles for enrichment. Once the tumour is developed and surgery is necessary to save the patient's life, a surgical guidance becomes important in order to assess the completeness of the removal of the cancerous tissue. Here, label-free and SERS-tag-based methods are developed as discussed in this review article. In the case of label-free approaches, sophisticated chemometrical tools are needed since the SERS spectra contain all information and are difficult to interpret. In order to simplify the SERS response, again SERS tags with specific cancer-related recognition units are of advantage. Here, the distribution of the marker band of the related reporter molecule is illustrated in order to identify the tumour margins. It is expected that the future lies on the application of SERS tags due to the ease of data interpretation and the accessibility to specific recognition units e.g. antibodies and aptamers, which are the centrepiece of the successful application of SERS tags. Based on the scientific achievements in the detection of cancer cells and tumour tissue, the next consequent step in bringing SERS toward clinical applications was to study the in vivo detection of cancer. In doing so, SERS tags showing excellent signal intensity are applied equipped with specific recognition elements allowing for a specific interaction of overexpressed cancer biomarkers. The Raman reporter molecules show a unique fingerprint spectrum, which is easily separated from the spectral background due to the complex biological matrix. Due to the tissue optical window allowing for penetration depths of several 100 μm into tissue within the NIR spectral range, the laser line at 785 nm is applied in most in vivo studies. Thus, SERS tags based on gold specifically designed for this wavelength are needed. The SERS method can be used for the tumour detection on the surface or underneath the skin; however, tumours buried in tissue cannot be monitored in 3D by means of the application of SERS tags. Therefore, SERS needs to be combined with imaging techniques such as magnetic resonance imaging if the 3D monitoring of cancer within the human body is aimed for. Moreover, in all studies the elimination processes of SERS tags from the organism have been neglected so far. When aiming for in vivo applications in the future, the nanotoxicity potential of those SERS tags needs to be considered as well as their fate in the organism. Studies should address where the SERS tags are accumulated and how the nanosensors can be eliminated e.g. via degradation processes. Thus, the design of SERS tags has to be reconsidered in agreement with safety by design concepts.
A further future topic of SERS in clinical diagnosis might be theranostics representing the combination of therapy and diagnostics. SERS is a perfect candidate as a tool in theranostic applications since the diagnostic SERS tags can be easily equipped with other modalities such as drug release and PTT as illustrated in the previous section. As an example, drug loaded nanoparticles are incorporated into cells and the drug release is monitored with SERS which has tremendous potential in pre-clinical research to test newly developed drugs. Moreover, new pathways of the drug into the cell might be opened when using nanostructures as the drug delivery vehicle. Based on the dramatic achievements of SERS in theranostics applications, a SERS-based theranostic platform is the most promising to pave the way for SERS from fundamental science to clinical applications by transferring the extraordinary scientific and technological advances into clinical practice. Next-generation theranostic tools urgently demand the integrated platform to enable rapid quantitative readout of target analytes in clinical samples with high specificity and sensitivity and also to implement therapy. Based on the improvement of the methodology, SERS-based multifunctional and multimodal platforms facilitate the comprehensive detection of biomarkers of disease which are effective as non-invasive, real-time surrogates for diagnostics and monitoring of therapeutic response, as well as providing therapeutic strategies via drug delivery or the photothermal effect.
Moreover, based on the development of techniques and instruments, it can be predicted that the most anticipated achievement in the future trend of SERS will be the development of integrative SERS image-guided on-site surgery platforms which will enable fast, ultrasensitive identification and removal of tumour primary sites and micrometastatic lesions during surgery processes via in vivo SERS imaging of tumours with high temporal–spatial resolution. To realize this great concept, cooperation and efforts are needed from the interdisciplinary fields of spectroscopy, biophotonics, materials science, analytical chemistry, and biomedical engineering.
Acknowledgements
Funding of the research projects “EXASENS” (13N13856) and “InfectoGnostics” (13GW0096F) by the Federal Ministry of Education and Research, Germany, is gratefully acknowledged. Furthermore, we thank the DFG for funding the research group FOR 1738 “Heme and heme degradation products”. Finally, the project MycoNET2 (01DN15028) is funded by the Federal Ministry of Education and Research, Germany, and the German Aerospace Center, which we gratefully acknowledge.References
- A. I. Henry, B. Sharma, M. F. Cardinal, D. Kurouski and R. P. Van Duyne, Anal. Chem., 2016, 88, 6638–6647 CrossRef CAS PubMed.
- D. Cialla, S. Pollok, C. Steinbrucker, K. Weber and J. Popp, Nanophotonics, 2014, 3, 383–411 CrossRef CAS.
- W. Xie and S. Schlucker, Rep. Prog. Phys., 2014, 77, 116502 CrossRef PubMed.
- K. A. Willets and R. P. Van Duyne, Annu. Rev. Phys. Chem., 2007, 58, 267–297 CrossRef CAS PubMed.
- S. Maier, Plasmonics: fundamentals and applications, Springer, New York, 2007 Search PubMed.
- M. Jahn, S. Patze, I. J. Hidi, R. Knipper, A. I. Radu, A. Muhlig, S. Yuksel, V. Peksa, K. Weber, T. Mayerhofer, D. Cialla-May and J. Popp, Analyst, 2016, 141, 756–793 RSC.
- A. Huefner, W. L. Kuan, K. H. Muller, J. N. Skepper, R. A. Barker and S. Mahajan, ACS Nano, 2016, 10, 307–316 CrossRef CAS PubMed.
- K. Bando, N. I. Smith, J. Ando, K. Fujita and S. Kawata, J. Opt., 2015, 17, 114023 CrossRef.
- F. Lussier, T. Brule, M. Vishwakarma, T. Das, J. P. Spatz and J. F. Masson, Nano Lett., 2016, 16, 3866–3871 CrossRef CAS PubMed.
- B. Kang, L. A. Austin and M. A. El-Sayed, ACS Nano, 2014, 8, 4883–4892 CrossRef CAS PubMed.
- S. R. Panikkanvalappil, S. M. Hira, M. A. Mahmoud and M. A. El-Sayed, J. Am. Chem. Soc., 2014, 136, 15961–15968 CrossRef CAS PubMed.
- S. R. Panikkanvalappil, S. M. Hira and M. A. El-Sayed, Chem. Sci., 2016, 7, 1133–1141 RSC.
- M. Aioub and M. A. El-Sayed, J. Am. Chem. Soc., 2016, 138, 1258–1264 CrossRef CAS PubMed.
- W. A. El-Said and J. W. Choi, Biotechnol. Bioprocess Eng., 2014, 19, 1069–1076 CrossRef CAS.
- W. A. El-Said, S. U. Kim and J. W. Choi, J. Mater. Chem. C, 2015, 3, 3848–3859 RSC.
- M. Xiao, L. Lin, Z. F. Li, J. Liu, S. L. Hong, Y. Y. Li, M. L. Zheng, X. M. Duan and X. Chen, Chem. – Asian J., 2014, 9, 2040–2044 CrossRef CAS PubMed.
- Y. L. Chen, L. Ding, W. Y. Song, M. Yang and H. X. Ju, Chem. Sci., 2016, 7, 569–574 RSC.
- W. Y. Song, L. Ding, Y. L. Chen and H. X. Ju, Chem. Commun., 2016, 52, 10640–10643 RSC.
- K. V. Kong, U. S. Dinish, W. K. O. Lau and M. Olivo, Biosens. Bioelectron., 2014, 54, 135–140 CrossRef CAS PubMed.
- P. Chen, Z. Y. Wang, S. F. Zong, H. Chen, D. Zhu, Y. Zhong and Y. P. Cui, Anal. Bioanal. Chem., 2014, 406, 6337–6346 CrossRef CAS PubMed.
- X. S. Zheng, P. Hu, Y. Cui, C. Zong, J. M. Feng, X. Wang and B. Ren, Anal. Chem., 2014, 86, 12250–12257 CrossRef CAS PubMed.
- Y. Cao, D. W. Li, L. J. Zhao, X. Y. Liu, X. M. Cao and Y. T. Long, Anal. Chem., 2015, 87, 9696–9701 CrossRef CAS PubMed.
- J. Cui, K. Hu, J. J. Sun, L. L. Qu and D. W. Li, Biosens. Bioelectron., 2016, 85, 324–330 CrossRef CAS PubMed.
- L. L. Qu, Y. Y. Liu, S. H. He, J. Q. Chen, Y. Liang and H. T. Li, Biosens. Bioelectron., 2016, 77, 292–298 CrossRef CAS PubMed.
- X. Gu, H. Wang, Z. D. Schultz and J. P. Camden, Anal. Chem., 2016, 88, 7191–7197 CrossRef CAS PubMed.
- W. A. El-Said, T. H. Kim, Y. H. Chung and J. W. Choi, Biomaterials, 2015, 40, 80–87 CrossRef CAS PubMed.
- J. Jiang, C. Auchinvole, K. Fisher and C. J. Campbell, Nanoscale, 2014, 6, 12104–12110 RSC.
- P. I. T. Thomson, V. L. Camus, Y. Y. Hu and C. J. Campbell, Anal. Chem., 2015, 87, 4719–4725 CrossRef CAS PubMed.
- Q. Liu, J. H. Wang, B. K. Wang, Z. Li, H. Huang, C. Z. Li, X. F. Yu and P. K. Chu, Biosens. Bioelectron., 2014, 54, 128–134 CrossRef CAS PubMed.
- A. Pallaoro, M. R. Hoonejani, G. B. Braun, C. D. Meinhart and M. Moskovits, ACS Nano, 2015, 9, 4328–4336 CrossRef CAS PubMed.
- X. X. Wu, Y. Z. Xia, Y. J. Huang, J. Li, H. M. Ruan, T. X. Chen, L. Q. Luo, Z. Y. Shen and A. G. Wu, ACS Appl. Mater. Interfaces, 2016, 8, 19928–19938 CAS.
- Z. A. Nima, M. Mahmood, Y. Xu, T. Mustafa, F. Watanabe, D. A. Nedosekin, M. A. Juratli, T. Fahmi, E. I. Galanzha, J. P. Nolan, A. G. Basnakian, V. P. Zharov and A. S. Biris, Sci. Rep., 2014, 4, 4752 CrossRef CAS PubMed.
- Z. F. Li, C. Li, D. Lin, Z. F. Huang, J. J. Pan, G. N. Chen, J. Q. Lin, N. R. Liu, Y. Yu, S. Y. Feng and R. Chen, Laser Phys. Lett., 2014, 11, 045602 CrossRef.
- C. M. Girish, S. Iyer, K. Thankappan, V. V. D. Rani, G. S. Gowd, D. Menon, S. Nair and M. Koyakutty, J. Mater. Chem. B, 2014, 2, 989–998 RSC.
- Y. Wang, S. Kang, A. Khan, G. Ruttner, S. Y. Leigh, M. Murray, S. Abeytunge, G. Peterson, M. Rajadhyaksha, S. Dintzis, S. Javid and J. T. C. Liu, Sci. Rep., 2016, 6, 21242 CrossRef CAS PubMed.
- L. Sinha, Y. Wang, C. Yang, A. Khan, J. G. Brankov, J. T. C. Liu and K. M. Tichauer, Sci. Rep., 2015, 5, 8582 CrossRef PubMed.
- J. K. Register, A. M. Fales, H. N. Wang, S. J. Norton, E. H. Cho, A. Boico, S. Pradhan, J. Kim, T. Schroeder, N. A. Wisniewski, B. Klitzman and T. Vo-Dinh, Anal. Bioanal. Chem., 2015, 407, 8215–8224 CrossRef CAS PubMed.
- A. D'Hollander, E. Mathieu, H. Jans, G. Vande Velde, T. Stakenborg, P. Van Dorpe, U. Himmelreich and L. Lagae, Int. J. Nanomed., 2016, 11, 3703–3714 CrossRef PubMed.
- R. J. Mallia, P. Z. McVeigh, C. J. Fisher, I. Veilleux and B. C. Wilson, Nanomedicine, 2015, 10, 89–101 CrossRef CAS PubMed.
- S. Jeong, Y. I. Kim, H. Kang, G. Kim, M. G. Cha, H. Chang, K. O. Jung, Y. H. Kim, B. H. Jun, D. W. Hwang, Y. S. Lee, H. Youn, K. W. Kang, D. S. Lee and D. H. Jeong, Sci. Rep., 2015, 5, 9455 CrossRef CAS PubMed.
- E. Garai, S. Sensarn, C. L. Zavaleta, N. O. Loewke, S. Rogalla, M. J. Mandella, S. A. Felt, S. Friedland, J. T. C. Liu, S. S. Gambhir and C. H. Contag, PLoS One, 2015, 10, e0123185 Search PubMed.
- Y. W. Wang, S. Kang, A. Khan, P. Q. Bao and J. T. C. Liu, Biomed. Opt. Express, 2015, 6, 3714–3723 CrossRef CAS PubMed.
- M. K. Hossain, H. Y. Cho, K. J. Kim and J. W. Choi, Biosens. Bioelectron., 2015, 71, 300–305 CrossRef CAS PubMed.
- L. J. Liang, D. S. Huang, H. L. Wang, H. B. Li, S. P. Xu, Y. X. Chang, H. Li, Y. W. Yang, C. Y. Liang and W. Q. Xu, Anal. Chem., 2015, 87, 2504–2510 CrossRef CAS PubMed.
- Y. Liu, J. R. Ashton, E. J. Moding, H. K. Yuan, J. K. Register, A. M. Fales, J. Choi, M. J. Whitley, X. G. Zhao, Y. Qi, Y. Ma, G. Vaidyanathan, M. R. Zalutsky, D. G. Kirsch, C. T. Badea and T. Vo-Dinh, Theranostics, 2015, 5, 946–960 CrossRef CAS PubMed.
- A. Carrouee, E. Allard-Vannier, S. Meme, F. Szeremeta, J. C. Beloeil and I. Chourpa, Anal. Chem., 2015, 87, 11233–11241 CrossRef CAS PubMed.
- W. Shi, R. J. Paproski, P. Shao, A. Forbrich, J. D. Lewis and R. J. Zemp, J. Biomed. Opt., 2016, 21, 020503 CrossRef PubMed.
- Y. Han, S. L. Lei, J. H. Lu, Y. He, Z. W. Chen, L. Ren and X. Zhou, Mater. Sci. Eng., C, 2016, 64, 199–207 CrossRef CAS PubMed.
- W. W. Zhang, Y. Q. Wang, X. Y. Sun, W. H. Wang and L. X. Chen, Nanoscale, 2014, 6, 14514–14522 RSC.
- T. Simon, M. Potara, A. M. Gabudean, E. Licarete, M. Banciu and S. Astilean, ACS Appl. Mater. Interfaces, 2015, 7, 16191–16201 CAS.
Footnote |
| † The authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |




