Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Real time detection of ESKAPE pathogens by a nitroreductase-triggered fluorescence turn-on probe†
Shengnan
Xu
a,
Qinghua
Wang
ab,
Qingyang
Zhang
ab,
Leilei
Zhang
b,
Limin
Zuo
c,
Jian-Dong
Jiang
ac and
Hai-Yu
Hu
 *ab
*ab
aState Key Laboratory of Bioactive Substances and Function of Natural Medicine, Institute of Materia Medica, Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing, 100050, China. E-mail: haiyu.hu@imm.ac.cn
bBeijing Key Laboratory of Active Substances Discovery and Drugability Evaluation, Institute of Materia Medica, Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing, 100050, China
cInstitute of Medicinal Biotechnology, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China
First published on 18th September 2017
Abstract
The identification of bacterial pathogens is the critical first step in conquering infection diseases. A novel turn-on fluorescent probe for the selective sensing of nitroreductase (NTR) activity and its initial applications in rapid, real-time detection and identification of ESKAPE pathogens have been reported.
Bacterial infections represent the leading cause of morbidity and mortality worldwide; especially, the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) cause the majority of hospital infections and effectively “escape” the effects of antibacterial drugs.1 In addition to novel treatment options, there is a strong need to better diagnose bacterial infections via non-invasive molecular imaging methods.2 To this end, various methods have been developed for the detection and identification of pathogens by nuclear imaging and optical imaging.3 Among these methods, fluorescence imaging has attracted much attention due to its high selectivity, good spatial resolution, and non-invasive mode of action.4 A number of bacteria targeting fluorescent probes have been developed in the past and several are on the way to the clinic.5 A common problem with these probes is the background fluorescent signal, which requires extensive and time-consuming washing steps to remove free fluorophores. Therefore, we hypothesized that bacteria specific enzyme-triggered fluorescence turn-on probes would provide a superior diagnostic capability for bacterial infections.6 These probes exhibit only low background fluorescence in a free state and, as a result, enable real time imaging of enzyme activities with a high signal-to-noise ratio in live bacterial cells. Furthermore, these probes also would be used for bacterial species identification via the differences of fluorescence turn-on response of each pathogen to the probe.
Nitroreductases (NTRs), a family of flavin-containing enzymes, are able to metabolize nitrosubstituted compounds using the reducing power of nicotinamide adenine dinucleotide (NAD(P)H).7 These enzymes are present in a wide range of bacterial genera and, to a lesser extent, in eukaryotes except hypoxic tumour cells and tissues, which make them a potential bacterial imaging target. Multiple sequence alignments of NTR proteins from E. coli and ESKAPE pathogens showed low similarity between each sequence pair (Fig. S1a, ESI†). Moreover, the phylogenetic tree constructed from amino acid sequences of NTR proteins also indicates that the NTRs of these seven species are not closely related (Fig. S1b, ESI†). Based on statistical analysis of sequences, we envisioned that it would be possible to diagnose ESKAPE pathogens by evaluating NTR activity detection.
The detection of NTR activities within hypoxic tumour cells and tissues using fluorogenic methods has been well described recently.8 In comparison, only a few optical probes have been developed for detecting NTR activities in E. coli and/or bacterial lysates,9 and there appears to have been no real time NTR enzymatic activity detected and evaluated in bacterial pathogens using a fluorogenic method. In an effort to detect NTRs which could be used for real time rapid detection and identification of ESKAPE pathogens, herein we designed a bacterial cell permeable fluorogenic probe which could selectively and rapidly be triggered by NTRs. Furthermore, the probe was successfully applied for imaging of NTRs in living bacterial cells and visualized the difference of NTR levels in ESKAPE pathogens by fluorescence confocal microscopy for the first time.
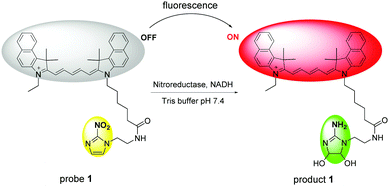
In our preliminary studies, we found that the lipophilic, near-infrared fluorescent cyanine dye Cy 5.5 with one positive charge could penetrate both Gram positive and Gram negative bacterial cell membranes and label ESKAPE pathogens (Fig. S2, ESI†); therefore, Cy 5.5 was chosen as a fluorochrome. The fluorogenic probe 1 was synthesized by the conjugation of a nitro-imidazole group (quencher) to Cy 5.5 and its structure was characterized by 1H NMR, 13C NMR and HRMS (see the ESI† for details). We expected the reduction of the nitro group into the amino group by NTR would result in an off–on fluorescence response (Scheme 1).
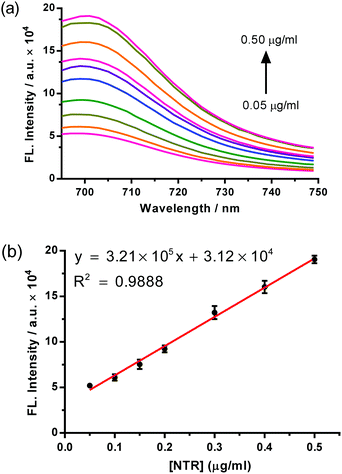
We first investigated the function response of probe 1 to purified E. coli NTR in vitro using fluorescence emission spectroscopy. Probe 1 was incubated in 0.05 M Tris buffer solution at pH 7.4, containing 500 μM NADH. As depicted in Fig. S3b (ESI†), the latent fluorescent probe 1 itself shows a very weak emission at 699 nm and the addition of NTR leads to an obvious fluorescence enhancement. The reduction product 1 was determined by high resolution mass spectrometry and confirmed by comparing with the chemically synthesized product 1 with HPLC (Fig. S4 and S15, ESI†) and the deduced process is proposed in Scheme S4 (ESI†).8a Furthermore, effects of pH and temperature on the reaction between probe 1 and NTR were studied (Fig. S5 and S6, ESI†), which reveals that the probe functions best under physiological conditions (about pH 7.4 and 37 °C). Under the optimized conditions, the fluorescence response of probe 1 to NTR at varied concentrations is shown in Fig. 1. A gradual increase in fluorescence intensity (FL intensity) was observed with the increase in the NTR concentrations, and a good linearity was obtained in the concentration range of 0.05–0.5 μg mL−1, with a linear equation of F = 3.21 × 105 C (μg per mL) + 3.12 × 104 (R = 0.9944). The detection limit (3σ/k, in which σ is the standard deviation of blank measurements, n = 11, and k is the slope of the linear equation) is determined to be 8.4 ng mL−1 NTR.
The apparent kinetic parameters of probe 1 catalysis by NTR were additionally determined. The rate of product 1 released from probe 1 triggered by NTR in the presence of NADH (500 μM) as an electron donor was measured by adding a known concentration of probe 1 (7.5–25 μM) to the reaction mixture. As shown in Fig. S7 (ESI†), fluorescence intensities increase with the level-up of the concentrations of probe 1 and nearly reach a plateau in about 30 min, which indicates that higher concentrations of probe 1 result in stronger fluorescence intensity. In contrast, the fluorescence of probe 1 without NTR (control) hardly alters during the same period of reaction time, also implying that the probe is stable in the detection system (Fig. S3b, ESI†).
Next, we investigated the kinetic parameters for the enzyme-activated reaction of probe 1. The Lineweaver–Burk plot of 1/V (V represents the initial reaction rate) versus the reciprocal of the probe concentration was established (Fig. S8, ESI†). By fitting the data with the Michaelis–Menten equation, the corresponding Michaelis constant (Km) and the maximum of the initial reaction rate (Vmax) for the enzyme-activated reaction were determined to be 7.21 μM and 0.020 μM s−1, respectively, which were comparable to the values reported previously.10
The specificity of probe 1 is extremely important when it is utilized for imaging NTR activity in bacteria. In order to explore the specificity of probe 1 to NTR, two sets of experiments based on interference factors and the NTR inhibitor were carried out. First, considering the complexity of the bacterial intracellular environment, the interferences of various bioanalytes were studied, including reductive biothiols (Cys, DTT, GSH and homocysteine), arginine (Arg), ascorbic acid (Vc), glucose, reactive oxygen species (ClO−, H2O2) and inorganic salts (CaCl2, MgCl2, KCl and NaCl). Fluorescence responses of probe 1 to the tested species showed that the fluorogenic probe 1 only could be triggered by NTR but not the other interference factors, including reductive biothiols at a high concentration (Fig. 2). These experiments provide evidence that probe 1 is highly specific to NTR and hence can be used for NTR detection.
Next, we investigated the effect of a common inhibitor of NTR, dicoumarin,11 on the activity of the enzyme. When probe 1 was pre-treated with 2.5 μM dicoumarin and then mixed with NTR, the fluorescence intensity was much lower than that without dicoumarin (Fig. S9, ESI†). Moreover, increasing the concentration of dicoumarin led to a progressive decrease in the fluorescence intensity of a mixture of probe 1 and NTR, confirming that the NTR activity was effectively inhibited by dicoumarin. These results confirmed that the enhanced fluorescence response of probe 1 to NTR arises from the enzyme-catalyzed reduction reaction.
Following the demonstration of the specificity of probe 1 to NTR, we examined whether probe 1 could be used to detect NTR activity in bacterial lysates. Grown E. coli cultures were treated with 5 μM of probe 1 at three different initial concentrations and subjected to subsequent lysis. As shown in Fig. 3, a gradual increase in fluorescence intensity is observed with the increase in the bacterial concentration. Moreover, the addition of dicoumarin (0.1 mM) to the bacterial culture before treatment with the probe significantly reduced the fluorescence intensity of the live E. coli, further demonstrating the specificity of the probe towards NTR.
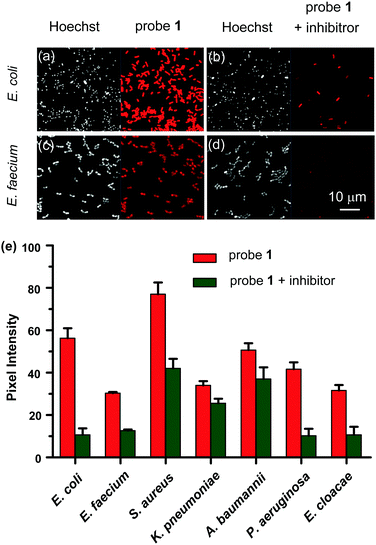
Encouraged by the in vitro imaging data, the ability of the NTR probe 1 for real time detection and identification of medically important bacterial pathogens of the ESKAPE panel, comprising the Gram negative species K. pneumoniae, A. baumannii, P. aeruginosa, E. cloacae and E. coli, and as well as the Gram positive species E. faecium and S. aureus, was probed. 5 μM probe 1 was incubated with seven bacterial strains in 0.05 M Tris buffer (pH 7.4) at 37 °C for 1 h, respectively, and the activation of probe 1 was analysed by confocal microscopy. As shown in Fig. 4(a–d) and Fig. S10 (ESI†), probe 1 was readily taken up and activated by all the seven tested bacterial cells. The intensity of the near-infrared fluorescence signal from Cy 5.5 indicated the relevant amount of probe 1 activated by NTRs in live bacteria. The quantification of Cy 5.5 fluorescence intensities in the cells revealed varying staining efficiencies among the ESKAPE pathogens (Fig. 4e). The strongest fluorescence signals from S. aureus incubated with probe 1 showed a 2.5-fold mean increase over the weakest one from E. faecium incubation. Meanwhile, pre-treatment of bacterial cells with the NTR inhibitor dicoumarin had a significant effect on probe 1 reduction, suggesting that the probe 1 was largely activated by NTR intracellularly after internalization. We believe the different fluorescence turn-on responses of probe 1 to ESKAPE pathogens are most likely due to the different amounts and/or types of NTRs in respective strains, which might make real time detection and identification of ESKAPE pathogens with NTR triggered probe 1 possible in the future. However, the exact molecular mechanism behind this finding remains unknown at this stage. To further evaluate the bacterial imaging efficiency of probe 1, we determined the time-dependent fluorescence responses of probe 1 in live methicillin-resistant S. aureus (MRSA). As depicted in Fig. S11a (ESI†), a visible fluorescence signal was observed in MRSA after an incubation time of 10 min and the Cy 5.5 fluorescence intensity reached to the highest value within approximately 1 h of incubation, suggesting probe 1 is capable of rapid detection of bacterial infections.
This work demonstrates the construction of a novel NTR triggered fluorescence turn-on probe 1 with potential applications in medical diagnostics for ESKAPE pathogens. Probe 1 displays a highly selective and rapid response of fluorescence enhancement to NTR. In particular, the probe, for the first time, has been successfully applied for real time rapid detection and comparative analysis of NTR activity in ESKAPE pathogens. Due to the substrate specificity of bacterial NTRs,12 further studies will focus on the design NTR activatable specific probes for rapidly identifying different pathogens within the ESKAPE collection.
We thank Prof. Ping Zhu, Dr Shu Xu and their group members (IMM) for their support and valuable discussions. This work was partially supported by the National Natural Science Foundation of China (NSFC) projects (21778077 and 21502236), Sino-German research project (GZ 1271) and the Beijing Nova Program (Z16111000490000).
Conflicts of interest
There are no conflicts to declare.Notes and references
- (a) M. Urdea, L. A. Penny, S. S. Olmsted, M. Y. Giovanni, P. Kaspar, A. Shepherd, P. Wilson, C. A. Dahl, S. Buchsbaum and G. Moeller, Nature, 2006, 444, 73–79 CrossRef PubMed; (b) L. B. Rice, J. Infect. Dis., 2008, 197, 1079–1081 CrossRef PubMed; (c) H. W. Boucher, G. H. Talbot, J. S. Bradley, J. E. Edwards, D. Gilbert, L. B. Rice, M. Scheld, B. Spellberg and J. Bartlett, Clin. Infect. Dis., 2009, 48, 1–12 CrossRef PubMed.
- M. van Oosten, M. Hahn, L. M. Crane, R. G. Pleijhuis, K. P. Francis, J. M. van Dijl and G. M. van Dam, FEMS Microbiol. Rev., 2015, 39, 892–916 CrossRef CAS PubMed.
- (a) A. Bunschoten, M. M. Welling, M. F. Termaat, M. Sathekge and F. W. van Leeuwen, Bioconjugate Chem., 2013, 24, 1971–1989 CrossRef CAS PubMed; (b) B. Mills, M. Bradley and K. Dhaliwal, Clin. Transl. Imaging., 2016, 4, 163–174 CrossRef PubMed; (c) Q. Zhang, Q. Wang, S. Xu, L. Zuo, X. You and H.-Y. Hu, Chem. Commun., 2017, 53, 1366–1369 RSC; (d) M. N. Pinto, I. Chakraborty, W. Schultz-Simonton, M. Rojas-Andrade, R. Braslau and P. K. Mascharak, Chem. Commun., 2017, 53, 1459–1462 RSC.
- (a) X. Li, X. Gao, W. Shi and H. Ma, Chem. Rev., 2013, 114, 590–659 CrossRef PubMed; (b) J. Wu, B. Kwon, W. Liu, E. V. Anslyn, P. Wang and J. S. Kim, Chem. Rev., 2015, 115, 7893–7943 CrossRef CAS PubMed.
- (a) X. Ning, S. Lee, Z. Wang, D. Kim, B. Stubblefield, E. Gilbert and N. Murthy, Nat. Mater., 2011, 10, 602–607 CrossRef CAS PubMed; (b) Y. Kong, H. Yao, H. Ren, S. Subbian, S. L. Cirillo, J. C. Sacchettini, J. Rao and J. D. Cirillo, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 12239–12244 CrossRef CAS PubMed; (c) M. van Oosten, T. Schafer, J. A. Gazendam, K. Ohlsen, E. Tsompanidou, M. C. de Goffau, H. J. Harmsen, L. M. Crane, E. Lim, K. P. Francis, L. Cheung, M. Olive, V. Ntziachristos, J. M. van Dijl and G. M. van Dam, Nat. Commun., 2013, 4, 2584 Search PubMed; (d) F. J. Hernandez, L. Huang, M. E. Olson, K. M. Powers, L. I. Hernandez, D. K. Meyerholz, D. R. Thedens, M. A. Behlke, A. R. Horswill and J. O. McNamara II, Nat. Med., 2014, 20, 301–306 CrossRef CAS PubMed; (e) G. Feng, Y. Yuan, H. Fang, R. Zhang, B. Xing, G. Zhang, D. Zhang and B. Liu, Chem. Commun., 2015, 51, 12490–12493 RSC; (f) K. Ferreira, H.-Y. Hu, V. Fetz, H. Prochnow, R. Rais, P. P. Müller and M. Brönstrup, Angew. Chem., Int. Ed., 2017, 56, 8272–8276 CrossRef CAS PubMed.
- (a) M. Kamiya, D. Asanuma, E. Kuranaga, A. Takeishi, M. Sakabe, M. Miura, T. Nagano and Y. Urano, J. Am. Chem. Soc., 2011, 133, 12960–12963 CrossRef CAS PubMed; (b) D. Asanuma, M. Sakabe, M. Kamiya, K. Yamamoto, J. Hiratake, M. Ogawa, N. Kosaka, P. L. Choyke, T. Nagano, H. Kobayashi and Y. Urano, Nat. Commun., 2015, 6, 6463 CrossRef CAS PubMed; (c) E. Calatrava-Perez, S. A. Bright, S. Achermann, C. Moylan, M. O. Senge, E. B. Veale, D. C. Williams, T. Gunnlaugsson and E. M. Scanlan, Chem. Commun., 2016, 52, 13086–13089 RSC; (d) S. Biswas, B. S. McCullough, E. S. Ma, D. LaJoie, C. W. Russell, D. Garrett Brown, J. L. Round, K. S. Ullman, M. A. Mulvey and A. M. Barrios, Chem. Commun., 2017, 53, 2233–2236 RSC; (e) X. He, L. Li, Y. Fang, W. Shi, X. Li and H. Ma, Chem. Sci., 2017, 8, 3479–3483 RSC; (f) Q. Zhang, Q. Wang, Y. Sun, L. Zuo, V. Fetz and H.-Y. Hu, Org. Lett., 2017, 19, 4496–4499 CrossRef CAS PubMed.
- (a) P. F. Searle, M. J. Chen, L. Hu, P. R. Race, A. L. Lovering, J. I. Grove, C. Guise, M. Jaberipour, N. D. James and V. Mautner, Clin. Exp. Pharmacol. Physiol., 2004, 31, 811–816 CrossRef CAS PubMed; (b) B. D. Palmer, P. van Zijl, W. A. Denny and W. R. Wilson, J. Med. Chem., 1995, 38, 1229–1241 CrossRef CAS PubMed; (c) C. Bryant and M. Deluca, J. Biol. Chem., 1991, 266, 4119–4125 CAS; (d) D. Bryant, D. McCalla, M. Leeksma and P. Laneuville, Can. J. Microbiol., 1981, 27, 81–86 CrossRef CAS PubMed.
- (a) R. B. Elmes, Chem. Commun., 2016, 52, 8935–8956 RSC; (b) X. You, L. Li, X. Li, H. Ma, G. Zhang and D. Zhang, Chem. – Asian J., 2016, 11, 2918–2923 CrossRef CAS PubMed; (c) Y. Zhou, K. N. Bobba, X. W. Lv, D. Yang, N. Velusamy, J. F. Zhang and S. Bhuniya, Analyst, 2017, 142, 345–350 RSC; (d) D. Li, Y. Xu, N. Zhou, J. Liu, R. Wang, T. Cheng, Y. Tang, W. Zhu, Y. Xu and X. Qian, Dyes Pigm., 2017, 136, 627–632 CrossRef CAS; (e) Z. R. Liu, Y. Tang, A. Xu and W. Lin, Biosens. Bioelectron., 2017, 89, 853–858 CrossRef CAS PubMed.
- (a) Z. Li, X. Gao, W. Shi, X. Li and H. Ma, Chem. Commun., 2013, 49, 5859–5861 RSC; (b) R. H. Wong, T. Kwong, K. H. Yau and H. Y. Au-Yeung, Chem. Commun., 2015, 51, 4440–4442 RSC.
- H. Nivinskas, R. L. Koder, Z. Anusevicius, J. Sarlauskas, A.-F. Miller and N. Cenas, Acta Biochim. Pol., 1999, 47, 941–949 Search PubMed.
- R. L. Koder and A.-F. Miller, Biochim. Biophys. Acta, 1998, 1387, 395–405 CrossRef CAS.
- M. D. Roldan, E. Perez-Reinado, F. Castillo and C. Moreno-Vivian, FEMS Microbiol. Rev., 2008, 32, 474–500 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Synthetic procedures and spectra data for all new compounds, and statistical analysis of NTR sequences, UV-VIS, FI spectral studies and confocal imaging studies. See DOI: 10.1039/c7cc07050k |
| This journal is © The Royal Society of Chemistry 2017 |