Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Impact of cellular health conditions on the protein folding state in mammalian cells†
Kohsuke
Inomata
 *ab,
Hajime
Kamoshida
c,
Masaomi
Ikari
a,
Yutaka
Ito
*ab,
Hajime
Kamoshida
c,
Masaomi
Ikari
a,
Yutaka
Ito
 c and
Takanori
Kigawa
c and
Takanori
Kigawa
 *ad
*ad
aRIKEN Quantitative Biology Center, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama 230-0045, Japan. E-mail: kinomata@riken.jp; kigawa@riken.jp
bPRESTO/Japan Science and Technology Agency, 4-1-8 Honcho, Kawaguchi, Saitama 332-0012, Japan
cDepartment of Chemistry, Graduate School of Science and Engineering, Tokyo Metropolitan University, 1-1 Minami-Osawa, Hachioji, Tokyo 192-0397, Japan
dDepartment of Computer Science, School of Computing, Tokyo Institute of Technology, 4259 Nagatsuta-cho, Midori-ku, Yokohama, Kanagawa 226-8503, Japan
First published on 22nd September 2017
Abstract
By using in-cell NMR experiments, we have demonstrated that the protein folding state in cells is significantly influenced by the cellular health conditions. hAK1 was denatured in cells under stressful culture conditions, while it remained functional and properly folded in cells continuously supplied with a fresh medium.
The cellular interior is a highly crowded and heterogeneous environment filled with various biomolecules, such as proteins, DNA, RNA, and other small metabolites.1 Since protein structures, dynamics, and activities can be influenced by intracellular viscosity, molecular crowding, and specific or non-specific interactions with biomolecules,2–10 protein behaviors in cells may be different from those in a dilute, homogeneous solution. In-cell NMR spectroscopy11 is a promising non-invasive technique to investigate the 3D structures,12–14 interactions,15,16 dynamics,17–19 and stabilities20,21 of biomolecules at the atomic level in living prokaryotic and eukaryotic cells.
However, most in-cell NMR studies have the potential concern that the protein behaviors in cells can be influenced by cellular damage under the unregulated cell culture conditions. In a typical in-cell NMR experiment, cells exist in an anaerobic high-density suspension (approximately 1 × 108 cells per mL) within the sample tube, in which nutrients can be rapidly depleted. Thus, many of the cells die within a few hours, in most cases. Therefore, in-cell NMR experiments must be completed in a very short time, to avoid protein leakage from dead cells.22,23 Moreover, a recent study demonstrated that the stress-induced increase in the cytosolic Ca2+ concentration converted metal-binding proteins from the Mg2+-bound state to the Ca2+-bound state.24 This suggests that the cellular health conditions should be regulated during in-cell NMR experiments, in order to investigate proteins sensitive to changes in the cellular state, such as by an external stimulus or stress response. In order to overcome this issue, bioreactor systems have recently been developed to maintain appropriate cell culture conditions during the experiments, by continuously supplying a fresh medium, and applied to structural analyses of proteins in E. coli25 and mammalian cells.26 The bioreactor system for in-cell NMR studies is quite critical for promoting cell survival and eliminating contamination of leaked isotope-labeled proteins from the sample. However, the means by which this improvement contributes to the protein folding state at an atomic level remain enigmatic.
In this study, we investigated the correlation between the cellular health conditions and the protein folding state in living mammalian cells, by using in-cell NMR spectroscopy with the bioreactor system. As a probe, we selected human adenylate kinase 1 (hAK1), which is a cytosolic enzyme catalyzing the reversible phosphoryl transfer reaction of adenine nucleotides in cells.27 The structure of adenylate kinase, which is conserved from bacteria to human,28 is composed of three domains:29 the central CORE domain, the ATP LID domain (LID), and the nucleoside monophosphate binding domain (NMPbind). The large conformational changes by the opening and closing of the LID and NMPbind domains are caused by the adenine nucleotide binding during the enzymatic cycle.30,31 In other words, the folding state of hAK1 is associated directly or indirectly with the environmental conditions, such as the ATP concentration, ionic strength, pH, and redox state. Therefore, this enzyme was considered to be an appropriate structural probe of the cellular health conditions.
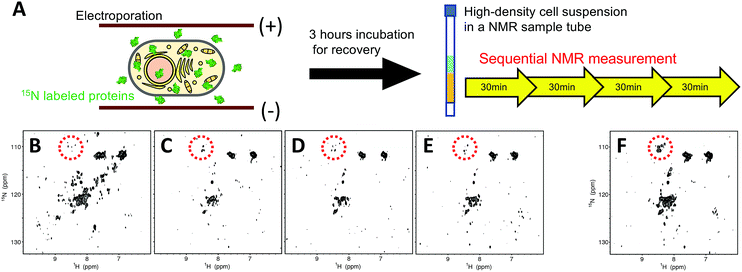
We first performed in-cell NMR experiments of hAK1 in a high-density HeLa cell suspension (approximately 1 × 108 cells per mL) without a fresh medium supply. The sample of HeLa cells containing 15N-labeled-hAK1 was prepared using the electroporation technique21,32 under optimized conditions (Fig. 1A; see Materials and methods, ESI,† for details). We confirmed, using confocal laser scanning microscopy, that the delivered hAK1 proteins were homogeneously distributed in the cytosol (Fig. S1, ESI†). In order to investigate the folding state of hAK1 in cells under these stressful conditions, we traced the time-dependent change of the 2D 1H–15N SOFAST-HMQC33 spectra (Fig. 1B–E). The hAK1 spectrum appeared to be relatively well dispersed at the start point; however, it became poorly dispersed after 30 minutes. The merger of three consecutive spectra (Fig. 1C–E) revealed broadened cross-peaks appearing around the 1H and 15N chemical shifts of 8.2–8.6 and 108–112 ppm, respectively (Fig. 1F), which is a characteristic pattern of partially or fully unfolded proteins.34,35 These results suggest that the deterioration in the cellular health conditions due to the unregulated cell culture can be related to the spectrum change into a poorly dispersed pattern, that is, the proteins unfolded during the in-cell NMR experiments.
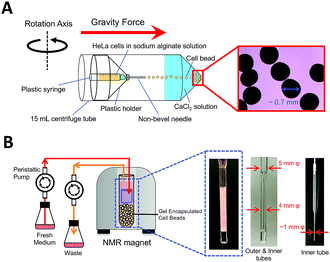
We expected that hAK1 could maintain the folded conformation in HeLa cells by avoiding the stressful culture conditions with the use of the bioreactor system. It is indeed an excellent method for in-cell NMR experiments. However, it was difficult to reproduce the method as reported, especially with respect to the cell immobilization by gel encapsulation, which required high technical skill or an additional apparatus. Therefore, we employed the centrifuge-based alginate gel encapsulation method36 for cell immobilization (Fig. 2A). The device was composed of commercially available components: a 15 mL centrifuge tube, a 1 mL syringe, a non-bevel needle, and a plastic holder. This method allowed us to easily obtain cell beads with highly reproducible sizes and distributions. Fig. 2B illustrates the schematic overview of our bioreactor system for in-cell NMR experiments. Gel encapsulated HeLa cell beads were packed into the custom-built NMR tube, and a culture medium was supplied to the HeLa cell beads from the bottom, through the tip of the custom-built inner tube made from a Pasteur pipette.
We evaluated our bioreactor system by performing an in-cell NMR experiment with a 15N-labeled human ubiquitin derivative harboring three alanine mutations (Leu8, Ile44 and Val70, designated as hUB-3A)20 in HeLa cells. A previous study demonstrated that the hUB-3A delivered into HeLa cells via the cell-penetrating-peptide mediated technique was homogenously distributed in the cytosol of the cells, resulting in good in-cell NMR spectra. We confirmed that the hUB-3A delivered by electroporation into HeLa cells was distributed similarly to the previous study (Fig. S2, ESI†) and it exhibited the most fully resolved in-cell NMR spectrum (Fig. S3, ESI†), demonstrating that our bioreactor system is applicable to in-cell NMR experiments.
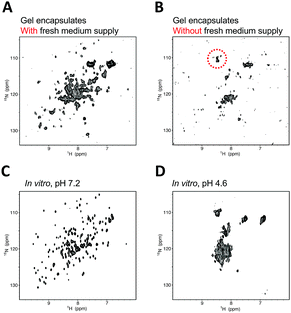
The hAK1 in HeLa cells with our bioreactor system exhibited a relatively well-resolved NMR spectrum at least for 12 hours (Fig. 3A), although several cross-peaks disappeared by signal line broadening, as compared with the in vitro reference (Fig. 3C), demonstrating that hAK1 adopts a folded conformation in HeLa cells during the in-cell NMR experiment. In contrast, the cross-peak pattern of hAK1 was poorly resolved in gel encapsulated HeLa cell beads without a fresh medium supply and resembled that in a high-density cell suspension (Fig. 3B), indicating that the deterioration in the cellular health conditions is due to the unregulated cell culture induced significant spectral changes. Since hAK1 maintained the folded conformation in HeLa cells with our bioreactor system, as mentioned above (Fig. 3A), we hypothesized that a stress induced denaturation of hAK1 in HeLa cells occurs under the unregulated culture conditions due to a simple acid denaturation (Fig. 3D) or a combination of various physicochemical properties under the stressful cellular environment, for example, cytosolic acidification, quinary interactions of bio-macromolecules,37 and so on. hAK1 may translocate into lysosomes with a luminal pH between 4.5 and 5.0,38 possibly via the chaperone-mediated autophagy (CMA) mechanism.39 Most of CMA substrates reportedly contain a peptide sequence biochemically related to KFERQ (KFERQ-like motif), which is recognized by the heat shock protein Hsc70. In fact, hAK1 contains a KFERQ-like sequence (26EKIVQ30), which hUB-3A does not have in turn. Therefore, hAK1 translocation into lysosomes under the stressful cellular environment may be feasible. A combination of various physicochemical properties under the stressful cellular environment may also induce the denaturation of hAK1. In addition to cytosolic acidification (Fig. S3, ESI†), some denaturation agent is likely to contribute to cause the effect similar to urea denaturation, which could not be simply explained by the macromolecular crowding effect (Fig. S4, ESI†).
We subsequently investigated the folding state of hAK1 located in the cytosolic space in HeLa cells, by using the in-cell NMR with our bioreactor system. As demonstrated in the previous studies,30,31 hAK1 mainly adopts two conformations in vitro: the open substrate-free form (Fig. 4A) and the closed substrate-bound form (Fig. 4B). The cross-peak pattern of hAK1 in HeLa cells is basically the same as that in the ATP saturated state in vitro, although several in-cell NMR cross-peaks are broadened and a few of them exhibited different chemical shifts from the corresponding ones in either the ATP saturated or the unsaturated state (Fig. 4C and D). This indicates that the hAK1 in HeLa cells basically binds adenine nucleotides in the closed conformational state. The delivered hAK1 concentration in HeLa cells was estimated to be on the order of 50–60 μM, based on the NMR signal intensities of 15N-labeled-hAK1 in the cell lysate. The intracellular ATP concentration in mammalian cells40 is typically on the millimolar order, in large excess over the delivered hAK1 in HeLa cells, coinciding with our results. In contrast, a recent study on E. coli adenylate kinase (ecAK) demonstrated that it adopted the open substrate-free conformation in E. coli cells,41 although the intracellular ATP concentration in E. coli is similar to that in mammalian cells.42 This suggests that more free ATP exists in the cytosol of mammalian cells than in E. coli cells, because a recent study also demonstrated that ecAK can form the closed substrate-bound conformation by administering a ribosome inhibitor and increasing the free ATP concentration in E. coli cells.41 As mentioned above, some of in-cell NMR signals of hAK1 exhibit different chemical shifts from those in either the closed or the open conformational state in vitro (Fig. 4C and D). This suggests that the details of the substrate-bound conformation and/or physicochemical properties of ATP,10 namely the hAK1 activity such as enzymatic turnover, are different from those in the diluted solution.
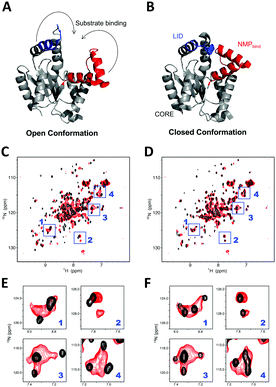 | ||
| Fig. 4 hAK1 conformation in HeLa cells with our bioreactor system. (A and B) Crystal structure of the substrate-free open AK1 (left, 3ADK.pdb) (A) and closed AK1 (right, 2C95.pdb) (B). LID (residues 131–141) is colored blue and NMPbind (residues 38–67) is colored red, respectively, and the arrows indicate the trajectories for closing LID and NMPbind into the closed conformation. (C and D) Superposition of in-cell NMR spectrum of 15N-labelled-hAK1 with our bioreactor system (red) and in vitro reference spectra of protein (black) in the absence (C) and presence (D) of ATP in excess. Blue boxes represent the regions with relatively large chemical shift differences between in the absence and presence of ATP in vitro. (E and F) Expansions of the boxed regions; (E) and (F) are derived from (C) and (D), respectively. | ||
In summary, we have demonstrated using in-cell NMR that the conformational state of hAK1 in HeLa cells is dramatically influenced by the cellular health conditions. By maintaining a continuous fresh medium supply with our bioreactor system, hAK1 remained in the functional, closed substrate-bound state in HeLa cells. The stressful culture conditions in the high-density cell suspension induced its conformational change into a partially or fully unfolded state. When investigating protein behaviors in cells by in-cell NMR, the proper regulation of the physiological conditions of the cells is crucial. This work provides a critical guide to in-cell NMR analyses, not only for protein structural analyses at an atomic level in mammalian cells but also for their potential applications in the future, such as drug candidate screening and molecular diagnoses of biopsy samples.
This work was supported in part by the Funding Program for the PRESTO program (no. JPMJPR13L2) and the Core Research for Evolutional Science and Technology (CREST) program (no. JPMJCR13M3) from the Japan Science and Technology Agency (JST), the AMED-SENTAN from Japan Agency for Medical Research and development (AMED), a Grant-in-Aid for Young Scientists B (no. JP25840061) and a Grant-in-Aid for Scientific Research on Innovative Areas (no. 25120003) from the Japan Society for the Promotion of Science (JSPS) and a RIKEN Pioneering Project, “Dynamic Structural Biology”. The authors thank Dr Hiromasa Yagi for helpful discussions and Ms Satoko Yasuda for secretarial assistance.
Conflicts of interest
There are no conflicts to declare.Notes and references
- O. Medalia, I. Weber, A. S. Frangakis, D. Nicastro, G. Gerisch and W. Baumeister, Science, 2002, 298, 1209–1213 CrossRef CAS PubMed.
- R. J. Ellis, Trends Biochem. Sci., 2001, 26, 597–604 CrossRef CAS PubMed.
- H.-X. Zhou, G. Rivas and A. P. Minton, Annu. Rev. Biophys., 2008, 37, 375–397 CrossRef CAS PubMed.
- K. Richter, M. Nessling and P. Lichter, Biochim. Biophys. Acta, Mol. Cell Res., 2008, 1783, 2100–2107 CrossRef CAS PubMed.
- Y. C. Kim, R. B. Best and J. Mittal, J. Chem. Phys., 2010, 133, 205101 CrossRef PubMed.
- A. H. Elcock, Curr. Opin. Struct. Biol., 2010, 20, 196–206 CrossRef CAS PubMed.
- M. Sarkar, C. Li and G. J. Pielak, Biophys. Rev., 2013, 5, 187–194 CrossRef PubMed.
- A. J. Wirth and M. Gruebele, BioEssays, 2013, 35, 984–993 CrossRef CAS PubMed.
- K. S. Hingorani and L. M. Gierasch, Curr. Opin. Struct. Biol., 2014, 24, 81–90 CrossRef CAS PubMed.
- I. Yu, T. Mori, T. Ando, R. Harada, J. Jung, Y. Sugita and M. Feig, eLife, 2016, 5, 1–22 Search PubMed.
- Z. Serber, A. T. Keatinge-Clay, R. Ledwidge, A. E. Kelly, S. M. Miller and V. Dötsch, J. Am. Chem. Soc., 2001, 123, 2446–2447 CrossRef CAS PubMed.
- D. Sakakibara, A. Sasaki, T. Ikeya, J. Hamatsu, T. Hanashima, M. Mishima, M. Yoshimasu, N. Hayashi, T. Mikawa, M. Wälchli, B. O. Smith, M. Shirakawa, P. Güntert and Y. Ito, Nature, 2009, 458, 102–105 CrossRef CAS PubMed.
- T. Ikeya, T. Hanashima, S. Hosoya, M. Shimazaki, S. Ikeda, M. Mishima, P. Güntert and Y. Ito, Sci. Rep., 2016, 6, 38312 CrossRef CAS PubMed.
- B.-B. Pan, F. Yang, Y. Ye, Q. Wu, C. Li, T. Huber and X.-C. Su, Chem. Commun., 2016, 52, 10237–10240 RSC.
- D. Burz, K. Dutta, D. Cowburn and A. Shekhtman, Nat. Methods, 2006, 3, 91–93 CrossRef CAS PubMed.
- L. Barbieri, E. Luchinat and L. Banci, Sci. Rep., 2015, 5, 14456 CrossRef CAS PubMed.
- Q. Wang, A. Zhuravleva and L. M. Gierasch, Biochemistry, 2011, 50, 9225–9236 CrossRef CAS PubMed.
- C. Li and M. Liu, FEBS Lett., 2013, 587, 1008–1011 CrossRef CAS PubMed.
- Y. Takaoka, Y. Kioi, A. Morito, J. Otani, K. Arita, E. Ashihara, M. Ariyoshi, H. Tochio, M. Shirakawa and I. Hamachi, Chem. Commun., 2013, 49, 2801 RSC.
- K. Inomata, A. Ohno, H. Tochio, S. Isogai, T. Tenno, I. Nakase, T. Takeuchi, S. Futaki, Y. Ito, H. Hiroaki and M. Shirakawa, Nature, 2009, 458, 106–109 CrossRef CAS PubMed.
- J. Danielsson, X. Mu, L. Lang, H. Wang, A. Binolfi, F.-X. Theillet, B. Bekei, D. T. Logan, P. Selenko, H. Wennerström and M. Oliveberg, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 12402–12407 CrossRef CAS PubMed.
- C. O. Barnes and G. J. Pielak, Proteins: Struct., Funct., Bioinf., 2011, 79, 347–351 CrossRef CAS PubMed.
- T. Sakai, H. Tochio, K. Inomata, Y. Sasaki, T. Tenno, T. Tanaka, T. Kokubo, H. Hiroaki and M. Shirakawa, Anal. Biochem., 2007, 371, 247–249 CrossRef CAS PubMed.
- D. S. S. Hembram, T. Haremaki, J. Hamatsu, J. Inoue, H. Kamoshida, T. Ikeya, M. Mishima, T. Mikawa, N. Hayashi, M. Shirakawa and Y. Ito, Biochem. Biophys. Res. Commun., 2013, 438, 653–659 CrossRef CAS PubMed.
- N. G. Sharaf, C. O. Barnes, L. M. Charlton, G. B. Young and G. J. Pielak, J. Magn. Reson., 2010, 202, 140–146 CrossRef CAS PubMed.
- S. Kubo, N. Nishida, Y. Udagawa, O. Takarada, S. Ogino and I. Shimada, Angew. Chem., Int. Ed., 2012, 51, 1–5 CrossRef.
- P. Dzeja and A. Terzic, Int. J. Mol. Sci., 2009, 10, 1729–1772 CrossRef CAS PubMed.
- T. T. Thach, T. T. Luong, S. Lee and D.-K. Rhee, FEBS Open Bio, 2014, 4, 672–682 CrossRef CAS PubMed.
- C. W. Müller, G. J. Schlauderer, J. Reinstein and G. E. Schulz, Structure, 1996, 4, 147–156 CrossRef.
- J. Adén and M. Wolf-Watz, J. Am. Chem. Soc., 2007, 129, 14003–14012 CrossRef PubMed.
- K. A. Henzler-Wildman, V. Thai, M. Lei, M. Ott, M. Wolf-Watz, T. Fenn, E. Pozharski, M. A. Wilson, G. A. Petsko, M. Karplus, C. G. Hübner and D. Kern, Nature, 2007, 450, 838–844 CrossRef CAS PubMed.
- Y. Hikone, G. Hirai, M. Mishima, K. Inomata, T. Ikeya, S. Arai, M. Shirakawa, M. Sodeoka and Y. Ito, J. Biomol. NMR, 2016, 66, 99–110 CrossRef CAS PubMed.
- P. Schanda and B. Brutscher, J. Am. Chem. Soc., 2005, 127, 8014–8015 CrossRef CAS PubMed.
- J. Balbach, V. Forge, W. S. Lau, N. A. van Nuland, K. Brew and C. M. Dobson, Science, 1996, 274, 1161–1163 CrossRef CAS PubMed.
- D. Eliezer, J. Yao, H. J. Dyson and P. E. Wright, Nat. Struct. Biol., 1998, 5, 148–155 CrossRef CAS PubMed.
- K. Maeda, H. Onoe, M. Takinoue and S. Takeuchi, Adv. Mater., 2012, 24, 1340–1346 CrossRef CAS PubMed.
- W. B. Monteith, R. D. Cohen, A. E. Smith, E. Guzman-Cisneros and G. J. Pielak, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 1739–1742 CrossRef CAS PubMed.
- J. A. Mindell, Annu. Rev. Physiol., 2012, 74, 69–86 CrossRef CAS PubMed.
- J. F. Dice, Autophagy, 2007, 3, 295–299 CrossRef CAS PubMed.
- A. Thore, S. Ansehn, A. Lundin and S. Bergman, J. Clin. Microbiol., 1975, 1, 1–8 CAS.
- S. Majumder, J. Xue, C. M. DeMott, S. Reverdatto, D. S. Burz and A. Shekhtman, Biochemistry, 2015, 54, 2727–2738 CrossRef CAS PubMed.
- H. Yaginuma, S. Kawai, K. V. Tabata, K. Tomiyama, A. Kakizuka, T. Komatsuzaki, H. Noji and H. Imamura, Sci. Rep., 2014, 4, 6522 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7cc06004a |
| This journal is © The Royal Society of Chemistry 2017 |