Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Automated glycan assembly of branched β-(1,3)-glucans to identify antibody epitopes†
M. W.
Weishaupt
ab,
H. S.
Hahm
 ab,
A.
Geissner
ab and
P. H.
Seeberger
ab,
A.
Geissner
ab and
P. H.
Seeberger
 *ab
*ab
aDepartment of Biomolecular Systems, Max Planck Institute of Colloids and Interfaces, Am Mühlenberg 1, 14476 Potsdam-Golm, Germany. E-mail: seeberger@mpikg.mpg.de
bDepartment of Chemistry and Biochemistry, Freie Universität Berlin, Arnimallee 22, 14195, Berlin, Germany
First published on 7th March 2017
Abstract
β-(1,3)-Glucans exhibit immunomodulatory and anti-tumor effects. Since the isolation of pure β-(1,3)-glucan oligosaccharides from natural sources is complicated, especially when certain branching patterns are desired, chemical synthesis is frequently the only means of accessing these molecules. We report the iterative automated glycan assembly (AGA) of conjugation-ready linear and branched β-(1,3)-glucan oligosaccharides.
β-(1,3)-Glucans are a heterogeneous class of carbohydrates found in fungi and bacteria.1,2 Fungal β-(1,3)-glucans can modulate the immune system of vertebrates3,4 and exhibit anti-tumor effects.5–7 Most fungal β-(1,3)-glucans also contain β-(1,6)-linked D-glucose side chains. The presence and distribution of β-(1,6) branching is essential for the immunomodulatory properties of this class of carbohydrates.8 The structural complexity of β-(1,3)-glucans may have a direct influence on these properties as well as their anti-tumor effects.9 β-Glucans are recognized by Dectin-1, a pattern recognition receptor of the innate immune system that belongs to the class of C-type lectin-like receptors.10–12 Binding of β-glucans to Dectin-1 induces phagocytosis and the production of proinflammatory factors by the innate immune system.13,14 Moreover, β-(1,3)-glucans significantly impact the adaptive immune response via activation of NFκB,15,16 as well as via antibody formation.17,18 Glucan structure has a strong impact on recognition by and interaction with recombinant murine Dectin-1.19 Studies based on isolated and possibly heterogeneous β-(1,3)-glucan samples may therefore lead to inconsistent and even contradictory reports.20–22 A collection of structurally defined β-(1,3)-glucans featuring a variety of different chain lengths and side chains would be helpful in determining the effect of length and branching pattern on antigenicity and receptor binding.
To date, longer β-glucan oligosaccharides have been synthesized exclusively by convergent solution phase syntheses.23–26 Convergent strategies limit the variability of accessible glucans regarding chain length, as well as position, number and length of branches. Utilizing an iterative automated solid-phase approach, we recently synthesized a linear β-(1,3)-glucan dodecasaccharide.27
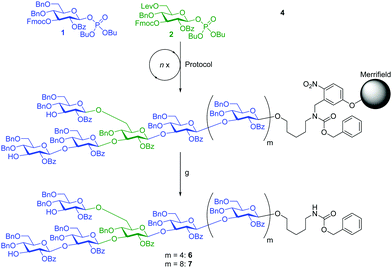
Here, we considerably improved and expanded this iterative strategy for fast and versatile access to β-(1,6) branched β-glucans, to prepare four β-(1,3)-glucan oligosaccharides that were subsequently immobilized for microarray based human antibody epitope evaluation. The iterative strategy for the assembly of branched β-(1,3)-glucans is based on building blocks 1 and 2 (Fig. 1).
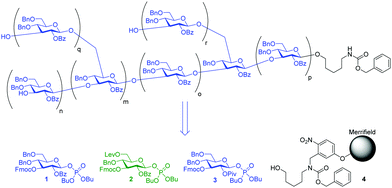 | ||
| Fig. 1 Variations in branching and chain length in β-(1,3)-glucans obtainable with building blocks 1 and 2 and solid support 4. | ||
The linear structure was assembled first, followed by the introduction of β-(1,6)-branching. Using this strategy, first the less nucleophilic secondary C3-hydroxyl groups are glycosylated before the reactive primary C6-hydroxyl groups serve as acceptors for branching. Glycosyl phosphate 3 (Fig. 1), originally developed for the synthesis of linear β-(1,3)-glucans,27 was modified by substituting the C2-pivaloyl with a benzoate ester in order to facilitate the final deprotection steps. The resulting building block 1 was used for linear assembly and also further modified to introduce β-(1,6)-linkages. Building block 2 contains a C6 levulinoyl ester that marks the branching points. Building blocks 1 and 2 were prepared applying standard protecting group chemistry (see ESI†) and enable the assembly of branched β-(1,3)-glucans of different chain length and side chain composition (Fig. 1).
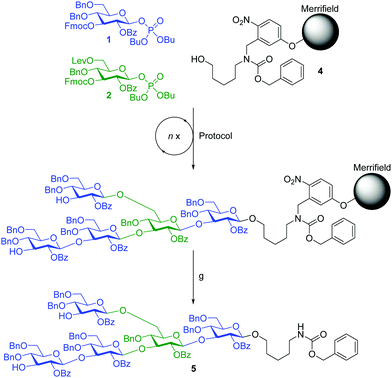
A previously described home-built oligosaccharide synthesizer28 was employed for all syntheses. Merrifield resin functionalized with photolabile linker 4 served as solid support for the incorporation of the monosaccharide building blocks.29 To avoid problems caused by residual base remaining after deprotection,27 an acidic washing step (TMSOTf in CH2Cl2 at −30 °C for 1 min) was introduced, preceding each glycosylation cycle and ensuring the removal of residual base and water after the previous deprotection step. The assembly of branched β-(1,3)-glucan pentasaccharide 5 served as a test for our automated glycan assembly (AGA) approach (Scheme 1).
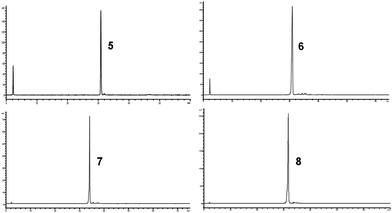
AGA of pentasaccharide 5 utilized three different glycosylation modules (A1, B1 and B1*, see Scheme 1 and ESI†). Each module uses three times three equivalents of glycosylating agent followed by an activator wash step with TMSOTf solution added at −30 °C, and the resin is agitated for 1 min before removal of the activator. Glycosylation module A1 activates three equivalents of glycosyl phosphate building block 1 with stoichiometric amounts of TMSOTf at −30 °C. After maintaining −30 °C for 10 min, the temperature is raised to −15 °C and maintained for 25 min. Glycosylation module B1 activates three equivalents of glycosyl phosphate building block 2 at −30 °C and allows 10 min for glycosylation, but raises the temperature to −10 °C and maintains it for 25 min to account for the lower reactivity of 2 caused by the presence of the Lev group. Glycosylation module B1* uses B1 reaction conditions for building block 1 since the C3-hydroxyl of 2 is expected to be a weaker nucleophile than its counterpart in building block 1. In all cases, the temporary Fmoc groups are removed with Et3N in DMF (10% v/v). The assembly of pentasaccharide 5 commenced with the reducing end glucose using module A1. For building block 2, module B1 was used. The glucose following the branching point was introduced using module B1*. The glucose residue at the non-reducing end was incorporated using module A1. Following selective removal of the Lev group at the branching position without cleavage of the Fmoc group at the non-reducing end glucose residue, the β-(1,6)-branch was introduced using module A1, and the two remaining Fmoc groups were removed. Following cleavage of the crude product from the resin by UV irradiation in flow,29 HPLC analysis showed one major peak (Fig. 2). Analysis by MALDI-TOF MS and NMR enabled identification of the desired branched β-(1,3)-glucan pentasaccharide 5 (ESI†). The correct installation of the desired 1,2-trans glycosidic linkages was confirmed by a coupled HSQC NMR experiment and by determining the C1–H1 coupling constants (JC1–H1 values between 161.1 and 162.5 Hz).
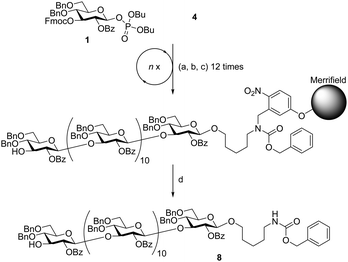
Branched β-(1,3)-glucan nonasaccharide 6 served as the next synthetic target (Scheme 2). In order to accelerate the synthesis, glycosylation modules A1, B1 and B1* were modified to provide two treatments with five equivalents of donor, leading to glycosylation modules A2, B2 and B2* (Scheme 2 and ESI†). The linear tetrasaccharide portion was assembled using glycosylation module A2 and standard Fmoc removal. Synthesis of the remaining pentasaccharide portion was performed by a sequence involving modules A2, B2, and B2* with standard Fmoc deprotection. The synthesis was completed using module A2, selective removal of the levulinoyl ester, activator wash, module A2, and final Fmoc deprotection. Analysis of the crude product by HPLC showed one major peak (Fig. 2). Isolation of that fraction and analysis by MALDI-TOF MS showed only the mass of the desired branched β-(1,3)-glucan nonasaccharide 6 (ESI†).
To expand on the approach, branched β-(1,3)-glucan tridecasaccharide 7 was assembled similar to nonasaccharide 6, with the difference that the linear portion of the structure was synthesized utilizing eight iterations of module A2 followed by Fmoc deprotection and activator wash (Scheme 2). Cleavage from the solid support afforded 120 mg crude product and, as before, analysis by HPLC showed one major peak (Fig. 2) and MALDI-TOF MS and NMR confirmed the identity of the desired tridecasaccharide 7 (ESI†).
In addition to branched structures 5–7, we also required linear dodecasaccharide β-(1,3)-glucan 8 to determine the effect of a side chain on anti-β-(1,3)-glucan antibody binding (Scheme 3). Cleavage from the solid support afforded 130 mg crude product with high purity (Fig. 2); analysis by MALDI-TOF MS and NMR revealed that the sole product of the synthesis was the desired linear β-(1,3)-glucan dodecasaccharide 8, and that plasticizer traces originating with the UV cleavage procedure were also present.
For the global deprotection of β-(1,3)-glucans 5–8, we first removed all benzoyl esters with NaOMe in CH2Cl2/MeOH. In the next step, all remaining protecting groups were removed by subjecting the compounds to Birch reduction conditions (Scheme 4). The crude products of the Birch reduction were purified over Sephadex® G-10 to afford fully deprotected and conjugation-ready β-(1,3)-glucan oligosaccharides 9, 10, 11 and 12.
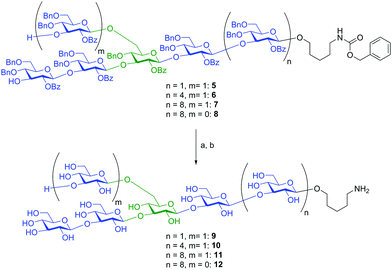 | ||
| Scheme 4 Global deprotection of β-(1,3)-glucan oligosaccharides 5, 6, 7 and 8. Reagents and conditions: (a) NaOMe, CH2Cl2/MeOH; (b) Na, NH3, t-BuOH, THF, (9: 21%; 10: 13%; 11: 60%; 12: 81%). | ||
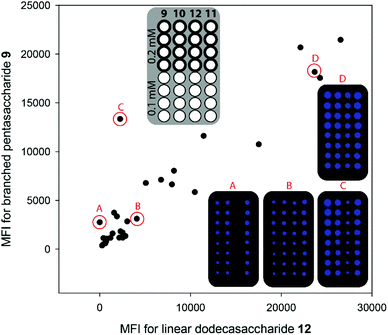
Antibodies against the linear β-(1,3)-backbone are thought to be protective while those against the β-(1,6)-branches are not.30 Antibodies against β-(1,3)-glucans have been described in the healthy human population, albeit at low and varying levels and no detailed epitope analysis is available.17,18 The oligosaccharides prepared by AGA allowed for analysis of these antibodies in whole serum. Glycan microarrays created by covalent immobilization of 9–12 on NHS activated glass slides were used to analyse sera from 31 healthy individuals.31,32 Briefly, the printed slides were incubated with sera, washed, and subsequent binding to an AlexaFluor labelled anti-human IgG-Fc enabled detection with a microarray scanner. Varying levels of IgG antibodies against β-(1,3)-glucans were observed in most of the tested individuals, with antibodies binding to both branched (9–11) and linear (12) glycans (Fig. 3 and Fig. S1, ESI†). As antibodies against the β-(1,6) branch are unlikely to bind to linear β-(1,3) dodecasaccharide 12, we can show that most individuals produce the protective antibody type. However, in some sera with low antibody levels the antibodies against the protective linear epitope were partially or completely diminished (individual A and cluster immediately below in Fig. 3). Although it is likely that antibodies that specifically recognize the β-(1,6) branch also exist in most individuals, the array data must not be overinterpreted because it cannot be excluded that even the smallest branched glycan, pentasaccharide 9, displays the linear epitope, as it contains a β-(1,3)-linked tetrasaccharide backbone. These microarrays show that the synthetic glycans prepared by AGA are a valuable tool to provide insights into β-glucan antibody specificity.
In summary, we describe the first iterative automated solid-phase synthesis of branched β-(1,3)-glucan oligosaccharides. This versatile synthetic route enables efficient access to a variety of conjugation-ready β-(1,3)-glucan oligosaccharides. The desired products were obtained without any deletion sequences and only one purification step following global deprotection was needed. Four β-(1,3)-glucan oligosaccharides were assembled and used to create glycan microarrays. Microarray analyses of human sera showed that most individuals form antibodies that bind to linear (protective) and branched β-(1,3)-glucan (non-protective) epitopes. This makes AGA an ideal tool for creating libraries of glucans for detailed studies of the natural antibody repertoire with respect to these important fungal cell surface antigens.
Notes and references
- B. A. Stone and A. E. Clarke, in Chemistry and Biology of (1 → 3)-[β]-glucans, ed. B. A. Stone and A. E. Clarke, La Trobe University Press, 1992 Search PubMed.
- A. E. Clarke and B. A. Stone, Biochim. Biophys. Acta, 1960, 44, 161 CrossRef CAS.
- G. C. Chan, W. K. Chan and D. M. Sze, J. Hematol. Oncol., 2009, 2, 25 CrossRef PubMed.
- M. Novak and V. Vetvicka, Endocr., Metab. Immune Disord.: Drug Targets, 2009, 9, 67 CrossRef CAS.
- K. Ina, T. Kataoka and T. Ando, Anti-Cancer Agents Med. Chem., 2013, 13, 681 CrossRef CAS PubMed.
- Y. Y. Maeda and G. Chihara, Nature, 1971, 229, 634 CrossRef CAS PubMed.
- Y. Y. Maeda, S. T. Watanabe, C. Chihara and M. Rokutanda, Cancer Res., 1988, 48, 671 CAS.
- A. Misaki, E. Kishida, M. Kakuta and K. Tabata, in Carbohydrates and Carbohydrate Polymers: Analysis, Biotechnology, Modification, Antiviral, Biomedical and Other Applications, ed. M. Yalpani, ATL Press, Science Publ, Inc., Mt Prospect, 1993, vol. 1, pp. 116–129 Search PubMed.
- J. A. Bohn and J. N. BeMiller, Carbohydr. Polym., 1995, 28, 3 CrossRef CAS.
- G. D. Brown and S. Gordon, Nature, 2001, 413, 36 CrossRef CAS PubMed.
- G. D. Brown, J. Herre, D. L. Williams, J. A. Willment, A. S. Marshall and S. Gordon, J. Exp. Med., 2003, 197, 1119 CrossRef CAS PubMed.
- P. Kougias, D. Wei, P. J. Rice, H. E. Ensley, J. Kalbfleisch, D. L. Williams and I. W. Browder, Infect. Immun., 2001, 69, 3933 CrossRef CAS PubMed.
- G. D. Brown, Nat. Rev. Immunol., 2006, 6, 33 CrossRef CAS PubMed.
- J. S. Schorey and C. Lawrence, Curr. Drug Targets, 2008, 9, 123 CrossRef CAS PubMed.
- B. A. Wevers, T. B. H. Geijtenbeek and S. I. Gringhuis, Future Microbiol., 2013, 8, 839 CrossRef CAS PubMed.
- S. I. Gringhuis, J. den Dunnen, M. Litjens, M. van der Vlist, B. Wevers, S. C. M. Bruijns and T. B. H. Geijtenbeek, Nat. Immunol., 2009, 10, 203 CrossRef CAS PubMed.
- P. Chiani, C. Bromuro, A. Cassone and A. Torosantucci, Vaccine, 2009, 27, 513 CrossRef CAS PubMed.
- I. Noss, I. M. Wouters, L. A. M. Smit, E. Meijer, A. Pronk, D. J. J. Heederik and G. Doekes, Int. Arch. Allergy Immunol., 2012, 157, 98 CrossRef CAS PubMed.
- E. L. Adams, P. J. Rice, B. Graves, H. E. Ensley, H. Yu, G. D. Brown, S. Gordon, M. A. Monteiro, E. Papp-Szabo, D. W. Lowman, T. D. Power, M. F. Wempe and D. L. Williams, J. Pharmacol. Exp. Ther., 2008, 325, 115 CrossRef CAS PubMed.
- B. H. Falch, T. Espevik, L. Ryan and B. T. Stokke, Carbohydr. Res., 2000, 329, 587 CrossRef CAS PubMed.
- K. Kataoka, T. Muta, S. Yamazaki and K. Takeshige, J. Biol. Chem., 2002, 277, 36825 CrossRef CAS PubMed.
- A. Mueller, J. Raptis, P. J. Rice, J. H. Kalbfleisch, R. D. Stout, H. E. Ensley, W. Browder and D. L. Williams, Glycobiology, 2000, 10, 339 CrossRef CAS PubMed.
- K. F. Mo, H. Q. Li, J. T. Mague and H. E. Ensley, Carbohydr. Res., 2009, 344, 439 CrossRef CAS PubMed.
- H. Tanaka, T. Kawai, Y. Adachi, N. Ohno and T. Takahashi, Chem. Commun., 2010, 46, 8249 RSC.
- G. Yang and F. Kong, Synlett, 2000, 1423 CAS.
- G. Yang and F. Kong, Carbohydr. Res., 2005, 340, 39 CrossRef CAS PubMed.
- M. W. Weishaupt, S. Matthies and P. H. Seeberger, Chem. – Eur. J., 2013, 19, 12497 CrossRef CAS PubMed.
- L. Krock, D. Esposito, B. Castagner, C.-C. Wang, P. Bindschadler and P. H. Seeberger, Chem. Sci., 2012, 3, 1617 RSC.
- S. Eller, M. Collot, J. Yin, H. S. Hahm and P. H. Seeberger, Angew. Chem., Int. Ed., 2013, 52, 5858 CrossRef CAS PubMed.
- C. Bromuro, M. Romano, P. Chiani, F. Berti, M. Tontini, D. Proietti, E. Mori, A. Torosantucci, P. Costantino, R. Rappuoli and A. Cassone, Vaccine, 2010, 28, 2615 CrossRef CAS PubMed.
- A. Geissner, C. Anish and P. H. Seeberger, Curr. Opin. Chem. Biol., 2014, 18, 38 CrossRef CAS PubMed.
- Healthy human sera were a kind gift of Dr Andreas Bergmann, Sphingotec GmbH, Hennigsdorf, Germany.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, Fig. S1, and full spectroscopic data for all new compounds. See DOI: 10.1039/c7cc00520b |
| This journal is © The Royal Society of Chemistry 2017 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: