Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Reversible DNA micro-patterning using the fluorous effect†
Gabriella E.
Flynn
 a,
Jamie M.
Withers
b,
Gerard
Macias
a,
Justin R.
Sperling
a,
Sarah L.
Henry
a,
Jonathan M.
Cooper
a,
Glenn A.
Burley
a,
Jamie M.
Withers
b,
Gerard
Macias
a,
Justin R.
Sperling
a,
Sarah L.
Henry
a,
Jonathan M.
Cooper
a,
Glenn A.
Burley
 *b and
Alasdair W.
Clark
*a
*b and
Alasdair W.
Clark
*a
aBiomedical Engineering Research Division, School of Engineering, University of Glasgow, Rankine Building, Oakfield Avenue, Glasgow, UK. E-mail: Alasdair.clark@glasgow.ac.uk
bWestCHEM & Department of Pure & Applied Chemistry, University of Strathclyde, 295 Cathedral Street, Glasgow, G1 1XL, UK. E-mail: glenn.burley@strath.ac.uk
First published on 23rd February 2017
Abstract
We describe a new method for the immobilisation of DNA into defined patterns with sub-micron resolution, using the fluorous effect. The method is fully reversible via a simple solvent wash, allowing the patterning, regeneration and re-patterning of surfaces with no degradation in binding efficiency following multiple removal/attachment cycles of different DNA sequences.
Of central focus to many chemical studies is molecular surface patterning, due to the potential impact of this research in scientific fields ranging from biosensing and diagnostics to computing.1 Of particular interest is the patterning of biomolecules, more specifically DNA. This is due to its propensity for specific molecular recognition and its ability to self-assemble.2 As such, its unique structural and chemical properties have seen it employed in an increasingly diverse range of applications. From biological screening to materials assembly, the integration of micro- and nano-patterned DNA with solid supports is being used to develop novel devices that bridge the gap between organic and inorganic engineering.3–5 As a result, efficient and flexible surface immobilization chemistries are paramount to the future development of DNA-based technologies. These chemistries must adhere to a number of stringent requirements: accessibility of the surface-bound DNA strands; intact functionality of the DNA base-pairing mechanism; high density of attachment; low non-specific binding; and high array stability. However, many immobilisation strategies fail to meet these requirements.6 Additionally, they tend to be static in nature, providing no opportunity to modify the composition of the DNA pattern after initial immobilisation.7–9 This restricts the potential applications of these surfaces, particularly in biosensing and diagnostics, where such devices are limited to a single use.10 Therefore, the development of chemistries that enable reversible and rewritable patterning would provide a route towards more versatile systems and devices. To that end, reversible DNA patterning has been demonstrated as a means to create complex micro-arrays for reusable screening applications.11,12 Furthermore, dynamic DNA patterns could provide a route towards a surface-based implementation of the transformable nanoparticle systems, which has been demonstrated recently in solution, for applications in reconfigurable nanophotonics.13
Currently, reusable DNA microarray chips typically rely on a stripping mechanism to refresh the surfaces between each use.14 This denatures the double stranded DNA leaving only the covalently bound single strand on the surface, which is then available to bind once more to the target. This process has many benefits, including reduced cost and higher consistency between experiments.15 Typically, high temperature detergents are required to strip the surface, which are potentially detrimental to the integrity of the chip. Furthermore, this technique only allows for the same probes to be screened multiple times.14 As a result, investigations concerning the development of new re-writable DNA platforms have become more widespread, and have concentrated on the use of disulphide bonds as reversible anchors for DNA immobilisation, as these covalent linkages are capable of reversible cleavage.16,17 Although these methods offer strong binding, they require a large excess of reagent to achieve an efficient regeneration, and are often limited by surface degradation after multiple cycles (due to, for example, thiol oxidation on surfaces).17 In contrast, fluorous surfaces are chemically inert, and the non-covalent nature of the “fluorous effect” allows for the complete removal of surface-bound biomolecules using simple solvent washes.18 In this paper, we show for the first time the implementation of the fluorous effect for the reversible immobilisation of DNA micro-patterns on solid supports.
The fluorous effect refers to the observation that highly fluorinated or perfluorinated compounds have a tendency to exclude themselves from both aqueous and organic phases.19 The resulting effect of this phenomenon is the product of reduced London dispersion forces between per-fluorinated compounds as a consequence of the extremely low polarizability of the C–F bond. Molecular tagging with fluorous “ponytails” has the effect of drawing compounds into fluorous layers – a feature that has been widely exploited in synthetic chemistry to facilitate product purification.19 ”Fluorous affinity” tags have also been used as an effective means of immobilising carbohydrates and peptides on surfaces, and in screening protein-small molecule interactions.18–26 Existing examples employ mechanical means to introduce samples, relying upon printing, stamping, or direct spotting of fluorous-tagged compounds in order to create defined, two-dimensional microarrays of molecular information.21–26 The major advantages of this technology are the low sample volumes required and the ability for multiplexing, whereby multiple targets can be screened for on the same chip. However, a new printing/stamping step is required each time the surface is to be re-used. This is also true of the fabrication of many DNA microarray chips.27 As a consequence, these techniques are often slow, are difficult to scale-up, and may rely on expensive printing devices.
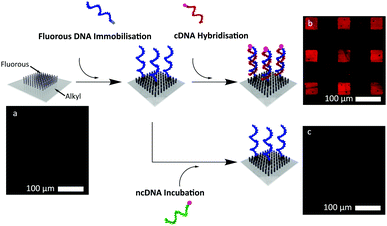
In this manuscript we demonstrate that (i) short fluorous tags immobilise water-soluble oligodeoxyribonucleotides (ODNs) from aqueous solutions onto lithographically-defined, micro-patterned fluorous surfaces without the need to direct sample spotting; (ii) ODNs immobilised by this method remain available to hybridise with a complementary ODN; and (iii) the genetic information can be completely removed using a simple organic solvent wash, and fully replaced with different genetic information during a short incubation step. This was done 5 times with no degradation of the surface throughout the subsequent immobilisations steps.
To demonstrate the specificity of the fluorous effect to immobilise ODNs into defined regions, without preventing the ability of the strands to participate in hybridisation events, we fabricated micro-patterned fluorous surfaces using standard optical lithography. These surfaces contained arrays of 50 × 50 μm squares modified with (heptadecafluoro-1,1,2,2-tetrahydrodecyl)trimethoxysilane, while the regions surrounding the squares were modified with n-decyltrichlorosilane (Fig. 1). A 1 μM solution of F-DNA1 (a 16-mer ODN containing an 8 carbon fluorous tag, see ESI†) was added to the surface and incubated for 2 hours before washing using TE buffer (10 mM TRIS, 1 mM EDTA (pH 8.0)). A 1 μM solution of the fluorescently labelled (TAMRA) complementary sequence, cDNA1, was then added to the surface and incubated in a humidity chamber for 2 hours. The surfaces were then rinsed in turn with TE buffer and DI water, and imaged using fluorescence microscopy at 20× magnification (0.4 NA). As Fig. 1b demonstrates, the fluorous immobilisation technique leaves the F-DNA1 strands available for hybridisation, and the F-DNA1/cDNA1 duplex was confined to the patterned fluorous areas.
One important issue in DNA microarray technology is non-specific binding, as this will ultimately limit both the sensitivity and the specificity of an assay.6 To test the extent of non-specific binding on our novel surfaces, we introduced a fluorescently labelled (TAMRA), non-complementary strand (ncDNA1) to a substrate modified with a fluorous-F-DNA1 micro-pattern. No fluorescence was observed upon addition of the non-complementary sequence to the surface, Fig. 1c, demonstrating that cDNA1 adhesion was mediated solely by hybridisation with the fluorous F-DNA1, see Fig. 1 and 2.
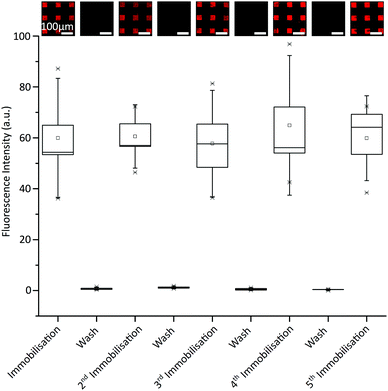
We next investigated the reversibility of the immobilisation and the durability of the fluorous surface over five DNA removal-replacement cycles. A range of solvents were screened to determine the optimal washing protocol, which was found to be a solution of 50% v/v MeOH in phosphate-buffered saline (PBS) (1 M, pH 7.4) followed by a methanol rinse. No loss of fluorescence intensity was observed after 5 cycles of immobilisation and washing with MeOH/PBS (Fig. 2).
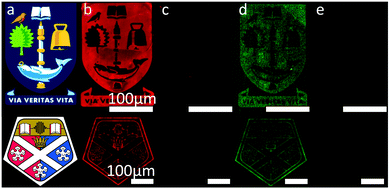
In order to assess if more complex information could be interchanged in place of less complex information on a surface, we synthesized a 32-mer ODN, F-DNA2, which contained a 5′fluorous tag and a complementary sequence, cDNA2, which contained an Alexa Fluor 488 tag. Further to this, we fabricated more intricate patterns using electron-beam lithography (Fig. 3), which contained feature sizes as small as 500 nm. Using our optimised conditions, described above, F-DNA1 was immobilised, then hybridised with its complement cDNA1 on the more intricate patterns. The duplex was then washed from the surface followed by the immobilisation of F-DNA2 and hybridisation with cDNA2. Here we see the potential of this immobilisation chemistry for applications in re-writable DNA microarray technology, where the same surface can be used multiple times to detect different targets. Further to this, in the case of the F-DNA2/cDNA2 duplex, the eight-carbon fluorous tag comprised less than 2% of the total mass. However, it was shown to be capable of immobilising the water-soluble ODNs with sufficient strength to remain bound to the surface through aqueous washes.
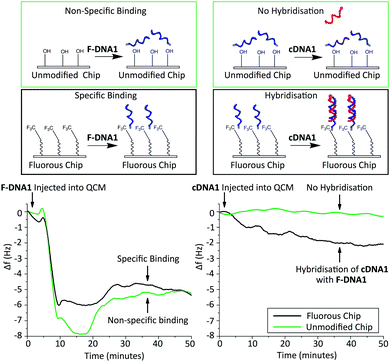
To obtain further understanding of the nature of the fluorous immobilisation, we monitored the binding events using a Quartz-Crystal Microbalance (QCM) (Fig. 4). This enabled us to investigate the binding events as a reduction in the resonance frequency of an oscillating 5 MHz crystal due to an increase in mass.28–30 To determine the specificity of the interaction, immobilisation of F-DNA1 was investigated using an unmodified silica QCM chip and a chip with a fluorous surface. Solutions of DNA (3.3 μM) were introduced to the chip using a flow cell at 40 μL min−1 and 20 °C. A decrease in frequency, representing an increase in surface mass, was observed upon addition of F-DNA1 and adsorption was complete within 30 minutes for both surfaces.31 Although F-DNA1 was adsorbed onto both fluorous and silica surfaces, after the introduction of the complementary sequence cDNA1, hybridisation was only observed in the case of F-DNA1 immobilised onto the fluorous QCM chip. In the case of F-DNA1 immobilised onto fluorous surfaces, the hydrophilic single-stranded DNA was directed away from the hydrophobic surface and toward the bulk solution, where it remained accessible to cDNA1. In the case where F-DNA1 was immobilised on silica, it is possible that charge-inversion of the silanol surface by cations present in the buffer results in the single-stranded DNA lying flat against the surface, where it was inaccessible to hybridisation.
In summary, we have demonstrated a novel method for the reversible immobilisation of DNA information onto surfaces. By developing lithographically patterned fluorous surfaces we remove the need to repeatedly direct the fluorous-modified biomolecules onto that surface for each subsequent “re-use”. The use of fluorous molecular tags enabled the immobilisation of ODNs to fluorous surfaces, and when used in conjunction with alkylated regions, allowed for the preparation of fluorescent patterns with low non-specific binding of the non-complementary strand to the sensing region and low non-specific binding of the target complementary strand to the non-sensing regions. ODNs immobilised using this method remained capable of hybridisation with complementary strands and displayed complete removal and replacement characteristics. This method enables the replacement of one DNA sequence for another, by a process that is repeatable and with no associated degradation of the efficiency of binding, with time. This reversible immobilisation chemistry could in the future be exploited across many research fields, particularly in DNA microarray development, where progress is already being made in fabricating re-usable sensing platforms.32
This work was supported by The EPSRC Centre for Doctoral Training (EP/F500424/1), the Royal Academy of Engineering (grant 10216/103), the EPSRC (grants EP/P51133X/1 and EP/N016874/1), and The Leverhulme Trust (grant RPG-2014-343). JC also acknowledges support from a personal EPSRC Fellowship (EP/K027611/1) and Biophononics ERC Advanced Investigator Award. The authors also wish to thank all the staff working in the James Watt Nanofabrication Centre for their support. All data relating to the work outlined in the article can be found here: http://dx.doi.org/10.5525/gla.researchdata.380.
Notes and references
- P. S. Cremer, J. Am. Chem. Soc., 2011, 133, 167–169 CrossRef CAS PubMed.
- A. M. Hung, H. Noh and J. N. Cha, Nanoscale, 2010, 2, 2530–2537 RSC.
- A. W. Clark and J. M. Cooper, Angew. Chem., Int. Ed., 2012, 51, 3562–3566 CrossRef CAS PubMed.
- A. W. Clark, D. G. Thompson, D. Graham and J. M. Cooper, Adv. Mater., 2014, 26, 4286–4292 CrossRef CAS PubMed.
- J. Zhang, S. Song, L. Wang, D. Pan and C. Fan, Nat. Protoc., 2007, 2, 2888–2895 CrossRef CAS PubMed.
- S. B. Nimse, K. Song, M. D. Sonawane, D. R. Sayyed and T. Kim, Sensors, 2014, 14, 22208–22229 CrossRef PubMed.
- D. I. Rozkiewicz, J. Gierlich, G. A. Burley, K. Gutsmiedl, T. Carell, B. J. Ravoo and D. N. Reinhoudt, ChemBioChem, 2007, 8, 1997–2002 CrossRef CAS PubMed.
- C. Wendeln, I. Singh, S. Rinnen, C. Schulz, H. F. Arlinghaus, G. A. Burley and B. J. Ravoo, Chem. Sci., 2012, 3, 2479–2484 RSC.
- I. Singh, C. Wendeln, A. W. Clark, J. M. Cooper, B. J. Ravoo and G. A. Burley, J. Am. Chem. Soc., 2013, 135, 3449–3457 CrossRef CAS PubMed.
- C. M. Niemeyer, L. Boldt, B. Ceyhan and D. Blohm, Anal. Biochem., 1999, 268, 54–63 CrossRef CAS PubMed.
- A. A. Lubin, R. Y. Lai, B. R. Baker, A. J. Heeger and K. W. Plaxco, Anal. Chem., 2006, 78, 5671–5677 CrossRef CAS PubMed.
- F. Huang, H. Xu, W. Tan and H. Liang, ACS Nano, 2014, 8, 6849–6855 CrossRef CAS PubMed.
- A. Kuzyk, R. Schreiber, H. Zhang, A. O. Govorov, T. Liedl and N. Liu, Nat. Mater., 2014, 13, 862–866 CrossRef CAS PubMed.
- X. Zhang, W. Xu, J. Tan and Y. Zeng, Anal. Biochem., 2009, 386, 222–227 CrossRef CAS PubMed.
- P. L. Dolan, Y. Wu, L. K. Ista, R. L. Metzenberg, M. A. Nelson and G. P. Lopez, Nucleic Acids Res., 2001, 29, e107–e107 CrossRef CAS PubMed.
- L. Li, W. Feng, A. Welle and P. A. Levkin, Angew. Chem., 2016, 128, 13969–13973 CrossRef.
- X. Du, J. Li, A. Welle, L. Li, W. Feng and P. A. Levkin, Adv. Mater., 2015, 27, 4997–5001 CrossRef CAS PubMed.
- R. L. Nicholson, M. L. Ladlow and D. R. Spring, Chem. Commun., 2007, 3906–3908 RSC.
- I. T. Horváth, D. P. Curran and J. A. Gladysz, in Handbook of Fluorous Chemistry, Wiley-VCH Verlag GmbH & Co. KGaA, 2005, pp. 1–4 DOI:10.1002/3527603905.ch1.
- C. M. Santos, A. Kumar, S. S. Kolar, R. Contreras-Caceres, A. McDermott and C. Cai, ACS Appl. Mater. Interfaces, 2013, 5, 12789–12793 CAS.
- H. D. Edwards, S. K. Nagappayya and N. L. B. Pohl, Chem. Commun., 2012, 48, 510–512 RSC.
- S. K. Mamidyala, K.-S. Ko, F. A. Jaipuri, G. Park and N. L. Pohl, J. Fluorine Chem., 2006, 127, 571–579 CrossRef CAS.
- H. D. Edwards, S. K. Nagappayya and N. L. B. Pohl, Chem. Commun., 2012, 48, 510–512 RSC.
- B. Y. M. Collet, T. Nagashima, M. S. Yu and N. L. B. Pohl, J. Fluorine Chem., 2009, 130, 1042–1048 CrossRef CAS PubMed.
- N. L. Pohl, Angew. Chem., Int. Ed., 2008, 47, 3868–3870 CrossRef CAS PubMed.
- G.-S. Chen and N. L. Pohl, Org. Lett., 2008, 10, 785–788 CrossRef CAS PubMed.
- M. Dufva, in DNA Microarrays for Biomedical Research: Methods and Protocols, ed. M. Dufva, Humana Press, Totowa, NJ, 2009, pp. 63–79 Search PubMed.
- F. Caruso, E. Rodda, D. N. Furlong, K. Niikura and Y. Okahata, Anal. Chem., 1997, 69, 2043–2049 CrossRef CAS PubMed.
- S. Yamaguchi, T. Shimomura, T. Tatsuma and N. Oyama, Anal. Chem., 1993, 65, 1925–1927 CrossRef CAS.
- Y. Okahata, Y. Matsunobu, K. Ijiro, M. Mukae, A. Murakami and K. Makino, J. Am. Chem. Soc., 1992, 114, 8299–8300 CrossRef CAS.
- G. Sauerbrey, Z. Phys., 1959, 155, 206–222 CrossRef CAS.
- A. A. Lubin, R. Y. Lai, B. R. Baker, A. J. Heeger and K. W. Plaxco, Anal. Chem., 2006, 78, 5671–5677 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures and further data. See DOI: 10.1039/c7cc00288b |
| This journal is © The Royal Society of Chemistry 2017 |