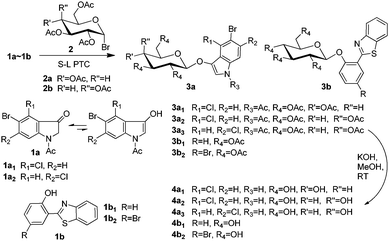
Synthesis of precipitating chromogenic/fluorogenic β-glucosidase/β-galactosidase substrates by a new method and their application in the visual detection of foodborne pathogenic bacteria†
Xianhu
Wei
abc,
Qingping
Wu
*b,
Jumei
Zhang
b,
Youxiong
Zhang
abc,
Weipeng
Guo
b,
Moutong
Chen
b,
Qihui
Gu
bd,
Zhihe
Cai
e and
Mianfei
Lu
e
aGuangzhou Institute of Chemistry, Chinese Academy of Sciences, Guangzhou 510650, China
bGuangdong Institute of Microbiology, State Key Laboratory of Applied Microbiology Southern China, Guangdong Provincial Key Laboratory of Microbial Culture Collection and Application, Guangdong Open Laboratory of Applied Microbiology, Guangzhou 510070, China. E-mail: wuqp203@163.com
cUniversity of Chinese Academy of Sciences, Beijing 100039, China
dSchool of Bioscience and Bioengineering, South China University of Technology, Guangzhou 510006, China
eGuangdong Huankai Microbial Sci. & Tech. Co., Ltd, Guangzhou 510663, China
First published on 18th November 2016
Abstract
We developed a new efficient method for the synthesis of important indoxyl glycoside substrates for β-glucosidase and β-galactosidase by using 1-acetylindol-3-ones as intermediates. This method was used to synthesise novel precipitating fluorogenic substrates for β-glucosidase based on 2-(benzothiazol-2′-yl)-phenols. We also assessed the application of these substrates in the detection of foodborne pathogenic bacteria.
The detection and identification of foodborne pathogenic bacteria are essential for evaluating food safety, an important public health concern worldwide.1 A new generation of agar medium assays based on synthetic enzymatic substrates, such as the most widely used tools, chromogenic agar media, has led to improved accuracy and faster detection compared with traditional culture medium. These assays are well suited for the differentiation and presumptive identification or screening of genera or species within polymicrobial cultures and can be easily integrated with other automated and/or molecular-based methods.2 For agar medium assays, the use of precipitating enzymatic substrates that produce insoluble signal products is necessary to avoid diffusion, which can hamper screening attempts. Among the precipitating enzymatic substrates, those for glycosidases, particularly β-glucosidase and β-galactosidase, are widely used and are typically based on indoxyls (indoxyl and its derivatives), namely indoxyl glycosides. Hydrolysis of indoxyl glycosides liberates a reactive indoxyl, which is oxidised by atmospheric oxygen to produce an insoluble dimeric and brightly coloured indigoid dye. For the indoxyl part, the two most common halide substitutions, 5-bromo-4-chloro- and 5-bromo-6-chloro-indoxyl (which are commonly abbreviated as X and Magenta, respectively), form a bright green/blue dye and a magenta dye, respectively.3
Although indoxyl glycosides are widely used in bacteriology, histochemistry, biochemistry, and molecular biology,3e the synthesis of these compounds is challenging. 1-Acetylindolin-3-ones are important intermediates, and glycosyl acceptors of these compounds have recently been obtained via a simple, robust method.4 However, improved 1-acetylindolin-3-one glycosylation with acetylated glycosyl halides, which is commonly carried out in acetone with sodium or potassium hydroxide with low yields, particularly for glucose derivatives, has not been reported. 3-Hydroxy-1H-indole-2-carboxylate has recently been applied; however, this method requires complex synthetic routes and produces low yields during decarboxylation of methyl ester derivatives.5
Compared with indoxyl glycosides, precipitating fluorogenic glycosides have not been well studied in microbiology.2,3b–d In recent years, various fluorescent probes based on 2-(benzothiazol-2′-yl)-phenols (BTPs), which exhibit excited-state intramolecular proton transfer (ESIPT; the enol form transforms reversibly into the keto form) upon irradiation to fluoresce more strongly and at longer wavelengths, have been developed for various applications.6 Precipitating fluorogenic glycosides, including cellulase substrates,7 galactosidase substrates,8 and sialidase substrates,9 which are based on BTPs, have also been reported for the screening or detection of valuable bacteria, purified β-galactosidase, and viruses and cancer. However, additional studies on the synthesis and application of such fluorogenic glycosides are still needed. Moreover, few studies have reported a combination of precipitating fluorogenic and chromogenic glycosides in bacteriology.
Herein, we develop a new method for the synthesis of indoxyl glycosides for β-glucosidase and β-galactosidase using easily available intermediates, i.e., 1-acetylindolin-3-ones, and novel precipitating fluorogenic glucosides based on BTPs (Scheme 1). We then investigated the combined application of these substrates.
Phase transfer catalysis (PTC) glycosylation was previously applied for the condensation of 1-acetyl-5-bromoindolin-3-one with peracetylated galactosyl bromide; however, the results were poor.10 After this, PTC glycosylation has been considered to be infeasible for 1-acetylindolin-3-ones.5c However, to our surprise, solid–liquid phase transfer catalysis (S–L PTC) glycosylation was found to be possible and feasible in our laboratory. A solid–liquid two-phase system including K2CO3, CHCl3, benzyltributylammonium chloride (BTBAC), and a small account of H2O was previously reported to be efficient for the preparation of glucosides of 2,6-dihydroxy-acetophenone and other substituted phenols.11 However, in our study, we found that the following reaction systems were efficient: tetrabutylammonium hydrogen sulphate (TBAHS) or tetrabutylammonium bromide (TBAB)/K3PO4/5% (w/w, based on the weight of the base) H2O/CH2Cl2/room temperature (RT, 25–28 °C) or reflux, and TBAHS/KOH/CH2Cl2/RT. Using our systems, the condensation of 1-acetyl-5-bromo-4-chloroindolin-3-one (1a1) with peracetylated glucosyl bromide (2a) could give the protected glycoside 3a1 with a yield of 46%, in which K3PO4 could outperform K2CO3 (40%) and Cs2CO3 (40%). However, 3.5% or 8% (w/w) H2O in the system TBAHS/K3PO4/CH2Cl2/RT caused a decrease in the yield of 3a1 to 38% or 29%, respectively. Additionally, for the TBAHS/KOH/CH2Cl2/RT system, if TBAHS was replaced with TBAB, then compound 3a1 could not be obtained. This might have been due to the presence of H2O, because TBAHS could react with KOH to generate a small amount of H2O, but TBAB could not. Additionally, an appropriate small amount of H2O could overcome the lattice energy of the solid base by the dissolution or formation of hydrated species, which then promote the occurrence of anion exchange and hence the initiation of the PTC process; but a further increase of H2O could progressively hamper the reaction.12 Because the strong base KOH yielded more black by-products, we preferred to use TBAHS/K3PO4/5% (w/w) H2O/CH2Cl2 for the preparation of other synthetic glycosides. Moreover, for glycosylation of 1-acetylindolin-3-ones with higher reactive glycosyl bromides, we preferred to carry out reactions at RT, and for the preparation of novel precipitating fluorogenic glucosides, we preferred to carry out reactions with refluxing. Consequently, the yields of the protected glycosides 3a2, 3a3, 3b1, and 3b2 under appropriate conditions (≤10 h) were 57%, 65%, 74%, and 90%, respectively.
From the above-mentioned results, we found that our S–L PTC method was feasible and efficient for glycosylation of 1-acetylindolin-3-ones and BTPs, particularly the latter. The hydroxyl groups of these types of molecules are restricted by keto forms or intramolecular hydrogen bonds, which makes glycosylation difficult. Moreover, we attempted several other methods for glycosylation of these types of molecules; however, these methods failed. For example, for the synthesis of 3a1 by the condensation of 1a1 and 2a, we also attempted to use the Koenigs–Knorr method7 and a reaction system containing NaH, tetrahydrofuran (THF), and dimethylformamide.8,9a However, these methods failed to yield 3a1. An improved Helferich glycosylation reaction using peracetyl sugars as glycosyl donors, developed in our laboratory for the efficient preparation of the protected fluorogenic 4-methylumbelliferyl glycoside substrates,13 was also attempted for the synthesis of 3a1 and 3b1, but failed to yield the expected products.
For deacetylation of the peracetyl glycosides, methanolysis using sodium methoxide (NaOMe) as a catalyst is often used.5e,5f,8 However, for deacetylation of 3a1, 3a2, 3a3, 3b1, and 3b2, we preferred methanolysis using KOH as a base catalyst, as previously reported,13 because NaOMe reacts readily with oxygen to form sodium formate and sodium hydroxide.14 Under our methanolysis conditions, the deacetylation of 3a1, 3a2, 3a3, 3b1, and 3b2 gave (5-bromo-4-chloroindol-3-yl) β-D-glucopyranoside (4a1, X-Glu), (5-bromo-4-chloroindol-3-yl) β-D-galactopyranoside (4a2, X-Gal), (5-bromo-6-chloroindol-3-yl) β-D-galactopyranoside (4a3, Magenta-Gal), [2-(benzothiazol-2′-yl)-phenolyl] β-D-glucopyranoside (4b1) and [2-(benzothiazol-2′-yl)-4-bromophenolyl] β-D-glucopyranoside (4b2) with yields of 89%, 86%, 99%, 99%, and 99%, respectively.
For application of the substrates, we first investigated the visual detection of purified β-glucosidase (β-Glu; 10 μg mL−1) activity using 4a1, 4b1, and 4b2 (80 μM each) under appropriate conditions (0.2 M AcOH–AcONa, pH 5.0, 37 °C, in air). After incubation of these substrates with active β-Glu (E+) for 45 min, 4a1 and 4b2 showed clear signal responses, whereas that of 4b1 was not observed until 90 min. For the respective control test groups using inactive β-Glu (E−) obtained by boiling treatment of E+ for 5 min, no significant changes were observed (Fig. S1, ESI†). These data demonstrated that 4a1, 4b1, and 4b2 could be used for the visual detection of β-Glu activity, but that 4a1 and 4b2 were more sensitive than 4b1. During the period of incubation with E+ for 90 min, followed by placement for 2 h at RT, 4a1 showed a light blue-green colour response under visible light and a relatively homogeneous strong blue fluorescence response under 365 nm ultraviolet (UV) light; this signal gradually disappeared after continuous placement at RT for 10.5 h (Fig. S1, ESI†). This showed that the relatively water-soluble X liberated by enzymatic hydrolysis of 4a1 was not readily oxidised by atmospheric oxygen to generate an insoluble blue-green indigoid dye in acidic medium. These data suggested that if 4a1 or other common substrates based on indoxyls are affected by acid-producing bacteria during application in the detection of foodborne pathogens, these substrates may show indigoid dye diffusion or weak indigoid dye responses. Because of the insolubility of fluorescent products 1b1 and 1b2 in water, the clear fluorescence responses of 4b1 and 4b2 were observed as many bright yellow-green, tiny dots in fluorescence emission. Additionally, these compounds, particularly 4b2, were gradually reduced to trace amounts, as shown by white turbidity or precipitation under visible light after incubation with E+ for 90 min followed by continuous incubation at RT (Fig. S1, ESI†).
Next, we evaluated the use of these probes in the visual detection of foodborne pathogenic bacteria. Common foodborne pathogenic bacteria, such as Salmonella, Escherichia coli, and Listeria monocytogenes, have been extensively investigated;15 however, foodborne Klebsiella pneumonia has not been reported frequently. This pathogen has been found in various foods from several countries, and antibiotic-resistant strains are commonly isolated from foods. Moreover, K. pneumonia has also been implicated as a causative foodborne pathogen in many diseases, such as ventilator-associated pneumonia, septicaemia, and various soft tissue infections.16 Herein, we performed the visual detection of K. pneumonia CMCC 46117 showing β-glucosidase activity using 4a1, 4b1, and 4b2 in different culture media under appropriate cultivation conditions (37 °C, in air). In nutrient broth containing 100 μM of substrate without negative bacteria, 4a1 showed a constant light blue-green colour response to the positive bacteria from 24 to 70 h of cultivation, whereas 4b1 and 4b2 showed no visible yellow-green fluorescence responses until 48 h of cultivation (Fig. S2, ESI†). Additionally, in Columbia agar containing 100 μM of substrate, with Salmonella typhimurium ATCC 14028 (showing no β-glucosidase activity) as a negative control, 4a1, 4b1, and 4b2 showed signal responses similar to those observed for positive bacteria alone within 24 h of cultivation (Fig. S3, ESI†). These results demonstrated that different media were associated with varying response efficiencies of the substrates. Moreover, all colonies of the positive bacteria on Columbia agar containing 4b1 or 4b2 were tightly localised, as shown by the yellow-green fluorescence signal, and were readily distinguished from the colonies of the negative control bacteria, which showed no fluorescence under UV light at 365 nm. Interestingly, the fluorescence from the colonies on Columbia agar containing 4b2 was much brighter. However, about half of the colonies of the positive bacteria on Columbia agar containing 4a1 were tightly localised, as shown by the blue-green indigoid dye signal, and the remaining colonies, particularly those surrounded by the negative bacteria, showed diffuse signals. Thus, these data demonstrated that 4b1 and 4b2 yielded better positional performance than 4a1.
Next, we performed simultaneously the visual detection of β-glucosidase and β-galactosidase activities of K. pneumonia CMCC 46117 as the positive bacteria on the same Columbia agar. The positive bacteria and Salmonella typhimurium ATCC 14028 (showing no β-glucosidase or β-galactosidase activities) as a negative control were inoculated together on different Columbia agar media containing two different substrates (100 μM + 100 μM and 0.06 g L−1 isopropyl-β-D-thiogalactoside [IPTG] for each agar) and cultivated for 24 h at 37 °C in air. The colonies of the positive bacteria on Columbia agar containing 4a1 and 4a3 were of blackish blue-green colour to the naked eye, a combination of blue-green and magenta under visible light, and no fluorescence under UV light at 365 nm; these colonies were not visibly different from most colonies of the positive bacteria on Columbia agar containing only 4a1 (Fig. S4, ESI†; Fig. 1A and B). However, all colonies of the positive bacteria on Columbia agar containing 4a2 and 4b2 or 4a3 and 4b2 were a clear blue-green or magenta colour under visible light and showed clear yellow-green fluorescence under UV light at 365 nm, except for a few small colonies or filament ribbon-like plaques, which were surrounded by the negative bacteria and showed a distinct colour but no visible fluorescence (Fig. S4, ESI†; Fig. 1C and D). These findings demonstrated that the respective response signals of 4b2 and the chromogenic substrates were mutually exclusive, and there were no significant mutual interferences during detection. Additionally, these findings showed that the combination of 4a2 or 4a3 and 4b2 could be used for the simultaneous visual detection of β-glucosidase and β-galactosidase activities from the same bacteria on the same agar, with an easy-to-distinguish and reliable signal, thereby improving the specificity and accuracy of the detection of the target bacterium. Moreover, the combination of blue-green and magenta responses could be readily influenced by varying the ratio of 4a1 to 4a3 and the ratio of their response efficiencies to respective glycosidase activities.
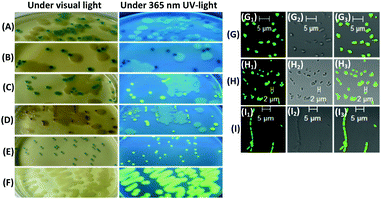 | ||
| Fig. 1 Responses of 4a1 (A), 4a1 + 4a3 (B), 4a2 + 4b2 (C), and 4a3 + 4b2 (D) to K. pneumonia CMCC 46117 and Salmonella typhimurium ATCC 14028; responses of 4a1 + 4b2 to Streptococcus faecalis ATCC 29212 (E) and B. cereus CMCC 63301 (F). Columbia agar plates were used, with 100 μM of each substrate, and cells were cultivated for 24 h at 37 °C in air. Partially enlarged images of Fig. S4 and S5 (ESI†). Confocal fluorescence images of colonies of K. pneumonia CMCC 46117 (G) from (D), Streptococcus faecalis ATCC 29212 (H) from (E) and B. cereus CMCC 63301 (I) from (F). (G1), (H1), and (I1) green emission (DAPI); (G2), (H2), and (I2) bright-field images; (G3), (H3), and (I3) merged images. λex = 405 nm. 40× magnification, NA: 1.30, oil immersion. Partially enlarged views of Fig. S6, ESI.† | ||
We also investigated the combination of the chromogenic substrate 4a1 and the fluorogenic substrate 4b2 for the visual detection of β-glucosidase activities of Streptococcus faecalis ATCC 29212 and Bacillus cereus CMCC 63301. After inoculation on Columbia agar containing 4a1 and 4b2 (100 μM each) and cultivated for 24 h at 37 °C in air, colonies of Streptococcus faecalis ATCC 29212 and B. cereus CMCC 63301 showed blue-green or no blue-green colour, respectively, under visible light and yellow-green fluorescence for both pathogens under UV light at 365 nm (Fig. S5, ESI†; Fig. 1E and F). Thus, it was not easy to generate significant mutual interference between 4b2 and any of the chromogenic substrates. Although 4a1 did not yield a signal response to B. cereus CMCC 63301, when 4a1 was combined with 4b2, the probability of detection of the target bacterium increased. Moreover, we could determine the relative activities of β-glucosidase, with the former being stronger than the latter.
For all the above experiments, the growth of the bacteria demonstrated that the novel substrates 4b1 and 4b2 were nontoxic or only slightly toxic to the four bacteria examined in this study.
β-Glucosidase herein was assumed to be an intracellular enzyme because the precipitating fluorescent compound 1b2 was limited to the colonies, and there were no fluorescent colonies surrounded by a fluorescence halo.17 This inference was further verified by examining the three bacteria from the brightly fluorescent colonies on Columbia agar containing 4a3 and 4b2 or 4a1 and 4b2 using laser confocal fluorescence microscopy. All three bacteria showed green emission (Fig. S6, ESI†; Fig. 1G–I), demonstrating that the hydrolysis of 4b2 occurred in bacterial cells and was catalysed by intracellular β-glucosidase, such that the bacteria were stained by the released fluorescent compound 1b2. Although 1b2 shows a maximum absorption peak at around 372 nm and a maximum fluorescence emission peak at around 526 nm,8 bacteria stained by this compound showed relatively clear green emission under 405 nm incident laser irradiation, using DAPI as a detection module. However, bacteria from the colonies showed fluorescence and blue-green or magenta colour was not observed to be visible under bright-field light using microscopy (Fig. S6, ESI†; Fig. 1G and H). Thus, these data demonstrated that 4b2 was more sensitive than 4a1 in bio-imaging experiments.
Considering that these probes are small molecules and similar to the corresponding natural glycose substrates, we speculate that their mechanism of ingestion into bacteria herein may also be a group translocation process via the phosphoenolpyruvate-dependent phosphotransferase system: the membrane-bound enzymes catalyze the transfer of the phosphoryl moiety of phosphoenolpyruvate to C-6 of the sugar moiety of the probe.18
In summary, we developed a new, efficient method for the synthesis of 4a1, 4a2, and 4a3 using easily available intermediates, 1-acetylindol-3-ones. This method was also much more efficient for the synthesis of 4b1 and 4b2 and involved solid–liquid phase transfer catalysis glycosylation, which can be carried out easily, with moderate to excellent yields (46–90%) within a relatively short reaction time (≤10 h). This method will also be beneficial for the synthesis of other glycosides, particularly novel glycosidase substrates, thereby promoting their application. Our findings herein, demonstrating the application of these substrates in the visual detection of K. pneumonia, Streptococcus faecalis and B. cereus, provide important advancements in the detection technology of foodborne pathogenic bacteria. In particular, they are very beneficial for advancements of chromogenic/fluorogenic agar media, paper-based analytical devices,19 and/or rapid enzyme assay techniques20 in microbiology.
We thank the National High Technology Research and Development Program (“863” Program) of China (2013AA102202), the Science & Technology Program of Guangdong Province (2012A080203014) and the Science & Technology Program of Guangzhou City (201604020003) for financial support.
References
- (a) M. Mangal, S. Bansal, S. K. Sharma and R. K. Gupta, Crit. Rev. Food Sci. Nutr., 2016, 56, 1568–1584 CrossRef PubMed; (b) Food Safety in Fact Sheets. Available online: http://www.who.int/mediacentre/factsheets/fs399/en/, accessed on 20 October 2016.
- (a) J. Merlino, Microbiology, 2010, 31, 127–130 Search PubMed; (b) E. L. Rank, Clin. Microbiol. Newsl., 2012, 34, 43–47 CrossRef; (c) E. L. Rank, Clin. Microbiol. Newsl., 2012, 34, 51–57 CrossRef.
- (a) J.-L. Reymond, V. S. Fluxa and N. Maillard, Chem. Commun., 2009, 34–46 CAS; (b) S. Orenga, A. L. James, M. Manafi, J. D. Perry and D. H. Pincus, J. Microbiol. Methods, 2009, 79, 139–155 CrossRef CAS PubMed; (c) M. Manafi, Int. J. Food Microbiol., 2000, 60, 205–218 CrossRef CAS PubMed; (d) J. Perry and A. Freydiere, J. Appl. Microbiol., 2007, 103, 2046–2055 CrossRef CAS PubMed; (e) J. A. Kiernan, Biotech. Histochem., 2007, 82, 73–103 CrossRef CAS PubMed; (f) H. M. Burke, T. Gunnlaugsson and E. M. Scanlan, Chem. Commun., 2015, 51, 10576–10588 RSC.
- M. N. Gandy, L. T. Byrne and K. A. Stubbs, Org. Biomol. Chem., 2015, 13, 905–908 CAS.
- (a) J. P. Horwitz, J. Chua, R. J. Curby, A. J. Tomson, M. A. Da Rooge, B. E. Fisher, J. Mauricio and I. Klundt, J. Med. Chem., 1964, 7, 574–575 CrossRef CAS PubMed; (b) Q. Wu, X. Wei, J. Zhang and W. Guo, Chemistry, 2013, 76, 580–589 CAS; (c) S. Böttcher and J. Thiem, Trends Carbohydr. Res., 2014, 6, 1–10 Search PubMed; (d) S. Böttcher, M. Hederos, E. Champion, G. Dékány and J. Thiem, Org. Lett., 2013, 15, 3766–3769 CrossRef PubMed; (e) S. Böttcher and J. Thiem, RSC Adv., 2014, 4, 10856–10861 RSC; (f) S. Böttcher and J. Thiem, Eur. J. Org. Chem., 2014, 564–574 CrossRef.
- (a) T. I. Kim, H. J. Kang, G. Han, S. J. Chung and Y. Kim, Chem. Commun., 2009, 5895–5897 RSC; (b) R. Hu, J. Feng, D. Hu, S. Wang, S. Li, Y. Li and G. Yang, Angew. Chem., Int. Ed., 2010, 49, 4915–4918 CrossRef CAS PubMed; (c) M. Santra, B. Roy and K. H. Ahn, Org. Lett., 2011, 13, 3422–3425 CrossRef CAS PubMed; (d) D. P. Murale, H. Kim, W. S. Choi and D. G. Churchill, Org. Lett., 2013, 15, 3946–3949 CrossRef CAS PubMed; (e) C. Huang, T. Jia, C. Yu, A. Zhang and N. Jia, Biosens. Bioelectron., 2015, 63, 513–518 CrossRef CAS PubMed; (f) Y. Jia, P. Li and K. Han, Chem. – Asian J., 2015, 10, 2444–2451 CrossRef CAS PubMed; (g) Z. M. Liu, L. Feng, G. B. Ge, X. Lv, J. Hou, Y. F. Cao, J. N. Cui and L. Yang, Biosens. Bioelectron., 2014, 57, 30–35 CrossRef CAS PubMed.
- D. R. Ivanen, N. L. Rongjina, S. M. Shishlyannikov, G. I. Litviakova, L. S. Isaeva-Ivanova, K. A. Shabalin and A. A. Kulminskaya, J. Microbiol. Methods, 2009, 76, 295–300 CrossRef CAS PubMed.
- T. Otsubo, A. Minami, H. Fujii, R. Taguchi, T. Takahashi, T. Suzuki, F. Teraoka and K. Ikeda, Bioorg. Med. Chem. Lett., 2013, 23, 2245–2249 CrossRef CAS PubMed.
- (a) A. Minami, T. Otsubo, D. Ieno, K. Ikeda, H. Kanazawa, K. Shimizu, K. Ohata, T. Yokochi, Y. Horii and H. Fukumoto, PLoS One, 2014, 9, e81941 Search PubMed; (b) Y. Kurebayashi, T. Takahashi, T. Otsubo, K. Ikeda, S. Takahashi, M. Takano, T. Agarikuchi, T. Sato, Y. Matsuda and A. Minami, Sci. Rep., 2014, 4 Search PubMed; (c) M. Takano, T. Takahashi, T. Agarikuchi, Y. Kurebayashi, A. Minami, T. Otsubo, K. Ikeda, H. Kanazawa and T. Suzuki, Virology, 2014, 464, 206–212 CrossRef PubMed.
- M. E. V. Dort, K. C. Lee, C. A. Hamilton, A. Rehemtulla and B. D. Ross, Mol. Imaging, 2008, 7, 187–197 Search PubMed.
- M. Hongu, K. Saito and K. Tsujihara, Synth. Commun., 1999, 29, 2775–2781 CrossRef CAS.
- D. Albanese, D. Landini, A. Maia and M. Penso, Ind. Eng. Chem. Res., 2001, 40, 2396–2401 CrossRef CAS.
- X. Wei, Y. Ma, Q. Wu, J. Zhang, Z. Cai and M. Lu, Molecules, 2015, 20, 21681–21699 CrossRef CAS PubMed.
- C. Barkenbus, V. C. Midkiff and R. M. Newman, J. Org. Chem., 1951, 16, 232–238 CrossRef CAS.
- (a) X. Yang, H. Li, Q. Wu, J. Zhang and L. Chen, Food Sci. Technol. Res., 2015, 21, 671–675 CrossRef CAS; (b) X. Yang, Q. Wu, J. Zhang, J. Huang, L. Chen, S. Liu, S. Yu and S. Cai, Food Control, 2015, 57, 308–313 CrossRef; (c) S. Zhang, Q. Wu, J. Zhang, Z. Lai and X. Zhu, Food Control, 2016, 68, 236–243 CrossRef CAS; (d) S. Zhang, Q. Wu, J. Zhang and X. Zhu, Foodborne Pathog. Dis., 2015, 13 Search PubMed; (e) M. Chen, Q. Wu, J. Zhang, S. Wu and W. Guo, Front. Microbiol., 2015, 6, 1026 Search PubMed; (f) S. Wu, Q. Wu, J. Zhang, M. Chen and W. Guo, Front. Microbiol., 2015, 7 Search PubMed.
- (a) E. Calbo and J. Garau, Clin. Infect. Dis., 2011, 52, 743–749 CrossRef PubMed; (b) R. K. Gautam, V. Nagar and R. Shashidhar, Radiat. Phys. Chem., 2015, 115, 107–111 CrossRef CAS; (c) H. S. Kim, J. W. Chon, Y. J. Kim, D. H. Kim, M. S. Kim and K. H. Seo, Int. J. Food Microbiol., 2015, 207, 83–86 CrossRef CAS PubMed; (d) W. Krusong, M. Teerarak and C. Laosinwattana, Food Control, 2015, 50, 502–508 CrossRef CAS; (e) Y. Guo, H. Zhou, L. Qin, Z. Pang, T. Qin, H. Ren, Z. Pan and J. Zhou, PLoS One, 2016, 11 Search PubMed.
- F. V. O. Kloeke, A. M. B. Iii, C. C. Eastburn, Z. Diwu and G. G. Geesey, J. Microbiol. Methods, 1999, 38, 25–31 CrossRef.
- (a) S. Roseman, J. Gen. Physiol., 1969, 54, 138–184 CAS; (b) S. S. Dills, A. Apperson, M. R. Schmidt and M. H. Saier Jr, Microbiol. Rev., 1980, 44, 385–418 CAS; (c) P. W. Postma, J. W. Lengeler and G. R. Jacobson, Microbiol. Rev., 1993, 57, 543–594 CAS; (d) J. Deutscher, C. Francke and P. W. Postma, Microbiol. Mol. Biol. Rev., 2006, 70, 939–1031 CrossRef CAS PubMed.
- J. C. Jokerst, J. A. Adkins, B. Bisha, M. M. Mentele, L. D. Goodridge and C. S. Henry, Anal. Chem., 2012, 84, 2900–2907 CrossRef CAS PubMed.
- L. Fiksdal and I. Tryland, Curr. Opin. Biotechnol., 2008, 19, 289–294 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6cc07522c |
| This journal is © The Royal Society of Chemistry 2017 |