Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Supramolecular surface functionalization via catechols for the improvement of cell–material interactions†
S.
Spaans
 ab,
P. P. K. H.
Fransen
ab,
P. P. K. H.
Fransen
 ac,
B. D.
Ippel
ac,
B. D.
Ippel
 ab,
D. F. A.
de Bont
ab,
H. M.
Keizer
d,
N. A. M.
Bax
ab,
C. V. C.
Bouten
ab and
P. Y. W.
Dankers
ab,
D. F. A.
de Bont
ab,
H. M.
Keizer
d,
N. A. M.
Bax
ab,
C. V. C.
Bouten
ab and
P. Y. W.
Dankers
 *abc
*abc
aInstitute for Complex Molecular Systems, Eindhoven University of Technology, De Zaale, 5612 AJ Eindhoven, The Netherlands. E-mail: p.y.w.dankers@tue.nl
bDepartment of Biomedical Engineering, Soft Tissue Engineering and Mechanobiology, Eindhoven University of Technology, P.O box 513, 5600 MB Eindhoven, The Netherlands
cDepartment of Biomedical Engineering, Laboratory of Chemical Biology, Eindhoven University of Technology, P.O. box 513, 5600 MB Eindhoven, The Netherlands
dSyMO-Chem BV, Eindhoven University of Technology, De Zaale, 5612 AZ Eindhoven, The Netherlands
First published on 13th June 2017
Abstract
Optimization of cell-material interactions is crucial for the success of synthetic biomaterials in guiding tissue regeneration. To do so, catechol chemistry is often used to introduce adhesiveness into biomaterials. Here, a supramolecular approach based on ureido-pyrimidinone (UPy) modified polymers is combined with catechol chemistry in order to achieve improved cellular adhesion onto supramolecular biomaterials. UPy-modified hydrophobic polymers with non-cell adhesive properties are developed that can be bioactivated via a modular approach using UPy-modified catechols. It is shown that successful formulation of the UPy-catechol additive with the UPy-polymer results in surfaces that induce cardiomyocyte progenitor cell adhesion, cell spreading, and preservation of cardiac specific extracellular matrix production. Hence, by functionalizing supramolecular surfaces with catechol functionalities, an adhesive supramolecular biomaterial is developed that allows for the possibility to contribute to biomaterial-based regeneration.
Introduction
Materials and systems inspired by nature have shown great promise for the development of cell adhesive surfaces and adhesive glues for biomedical applications.1–4 In the tissue engineering field researchers have been inspired by the adhesive properties of marine mussels. Mussels show superior adhesion on wet surfaces through their byssal threads.5–7 Within these threads, proteins containing the amino acid 3,4-dihydroxyphenylalanine (DOPA), are secreted in a spatially and temporally controlled manner which then form adhesive plaques in minutes.8 The strong adhesive properties of these threads originate from catechol residues of DOPA, which have efficient and versatile cross-linking abilities. In reducing conditions, catechol residues can undergo various interactions, such as hydrogen bonding, electrostatic interactions or metal-catechol coordination.9 Once exposed to oxygen, catechol residues are oxidized to reactive o-quinones. The o-quinones can then (i) self-polymerize, (ii) react with thiols or amines via Michael type addition or (iii) react with amines via Schiff base reactions.Many of these cross-linking abilities have contributed to the development of synthetic materials with the ultimate goal to promote adhesion of cells or biomolecules for tissue engineering applications.10,11 Catechol functionalities can be introduced by immersing material substrates in an alkaline solution of 3,4-dihydroxyphenethylamine (dopamine). Thereby a thin polydopamine film is formed that exhibits reactivity towards nucleophiles, such as thiols and amines.4,12,13 Polydopamine modified materials can also be used to immobilize extracellular matrix (ECM) proteins, such as collagen type I14 or serum proteins.13 To create polydopamine coatings on biomaterials, a large amount of dopamine is needed. To prevent the use of excessive dopamine, catechols can be directly coupled to synthetic polymers using dopamine or 3,4-dihydroxyhydrocinnamic acid, resulting in the formation of amide bonds. Brubaker et al. coupled catechols to a 4-arm poly(ethylene glycol) (PEG) and incorporated an elastase-specific peptide sequence to create an enzymatically degradable and adhesive hydrogel.15 Subcutaneous injection of this adhesive hydrogel in mice showed slow degradation via elastase secreted by neutrophils, which minimized an inflammatory response.15 Furthermore, Lee et al. conjugated catechol residues to the backbone of alginate, to develop biocompatible 3D hydrogel for cell transplantation.16 By exposing the catechol modified alginate to a mixture of sodium hydroxide and sodium periodate, a divalent cation-free alginate hydrogel was created that showed a high degree of viability and low immunogenicity. Kim et al. simultaneously electrospun catechol-modified 8-arm PEG with thiolated poly(lactic-co-glycolic acid) and either formed a cross-linked nanofibrous network after exposure to sodium periodate solution, or formed non cross-linked meshes without exposure to sodium periodate.17 Electrospun catechol cross-linked meshes showed improved anti-fouling properties compared to non-cross-linked meshes, due to the slower degradation rate of tethered PEG chains. Exemplary, Choi et al. covalently immobilized 3,4-dihydroxyhydrocinnamic acid to amines on the surface of polycaprolactone-PEG electrospun nanofibers.18 After exposure to basic pH, a low concentration of catechols was able to induce increased fibroblast adhesion at the surface of nanofibers (catechol concentration = 106 pmol mm−2). In general, these findings show an enhanced cellular adhesion attributed to the conjugation of catechol-molecules to polymers. Yet, since these methods require multiple reaction steps for functionalization of material surfaces, new approaches that limit the use of excessive conjugation reactions are highly desirable.
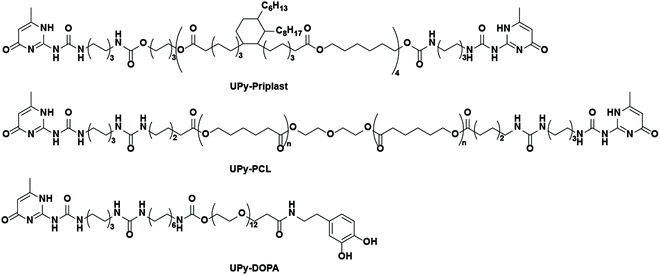
Therefore, here a supramolecular and modular approach is introduced based on supramolecular hydrogen bonding ureido-pyrimidinone (UPy) polymers and UPy-modified mussel-inspired catechol additives (abbreviated as UPy-DOPA), in order to develop cell-adhesive biomaterials (Scheme 1). For this purpose, first a new UPy-based hydrophobic polymer was developed, based on priplast, an amorphous polyester polyol. This UPy-priplast was proposed to display non-cell adhesive properties. Additionally, another, by our group developed and intensively studied, less hydrophobic, UPy-modified polyester UPy-polycaprolactone (UPy-PCL) was bioactivated with UPy-DOPA additives. We have been shown in the past that this UPy-PCL material is intrinsically cell adhesive (in the presence of serum proteins), and can therefore be applied as control material.19–22
As biological application and read-out we aimed at the improvement of cardiomyocyte progenitor cells (CMPC)-material interactions in terms of adhesion, viability and preservation of endogenous ECM formation. CMPC were chosen as cell source, because cardiac stem/progenitor cells have the potential to regenerate cardiac tissue and provide a long term solution for cardiac tissue damage when used as a cell-therapy.23,24 However, it remains a challenge to improve the engraftment and survival of these progenitor cells using biomaterials.
Materials & methods
Materials and synthetic procedures
Bifunctional UPy-modified polycaprolactone (UPy-PCL, Mn = 2.7 kg mol−1) was synthesized similar to previous methods.25 Bifunctional UPy-modified priplast3196 (UPy-priplast, Mn = 5.0 kg mol−1) was synthesized by reacting dihydroxyl-terminated priplast3196 with hexamethylene diisocyanate (HDI)-activated UPy using dibutyltin dilaurate (DBTDL) as a catalyst in CHCl3 (synthetic procedure and Scheme S1†). Monofunctionalized UPy-catechol (UPy-DOPA) was synthesized by coupling 3,4-dihydroxyphenethylamine to carboxylic acid terminated UPy-building blocks (Scheme S2†).26 General materials and instrumentations used in this study are described in the ESI.†Preparation of UPy-priplast films and tensile testing
Solutions of UPy-priplast in chloroform (15 mg mL−1) were stirred overnight, casted into Teflon molds (SyMo-Chem B.V., 4.5 × 10.5 × 0.5) and placed under a glass bowl to ensure slow evaporation of CHCl3. Dog bone shaped molds (18 × 5 mm) were used to cut out dog bone shaped polymer films. Stress–strain measurements were performed on a Zwick Z010 Tensile tester (Zwick Roell). Samples were placed in clamps with a crosshead of 18 mm and a pre-load of 0.1 N was applied with a speed of 0.3 mm s−1.27 Samples were loaded until break with a loading speed of 1 mm min−1. Strain-at-break, stress-at-break and Young's modulus was calculated from the loading curve using Matlab (Mathworks).Preparation and analysis of dropcast films
Polymer solutions were prepared by mixing UPy-PCL (31.8 mg ml−1 in HFIP) or UPy-Priplast (38.8 mg ml−1 in CHCl3) with UPy-DOPA (1.67 mg ml−1 in CHCl3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (95
MeOH (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5)), resulting in mixtures with 10 mol% UPy-DOPA. UPy-PCL and UPy-Priplast based films were prepared by dropcasting 40 μL and 80 μL polymer solutions on untreated glass coverslips and silanized glass coverslips (ESI 1.3†), respectively. Pure UPy-DOPA solutions (25 mg ml−1 in CHCl3
5)), resulting in mixtures with 10 mol% UPy-DOPA. UPy-PCL and UPy-Priplast based films were prepared by dropcasting 40 μL and 80 μL polymer solutions on untreated glass coverslips and silanized glass coverslips (ESI 1.3†), respectively. Pure UPy-DOPA solutions (25 mg ml−1 in CHCl3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (95
MeOH (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5)) were dropcast on silanized glass coverslips. Dropcast films were dried in vacuum at room temperature overnight. Fibronectin-coated cover glasses were prepared by incubating them in 30 μg mL−1 fibronectin in PBS for 1–2 hours. Atomic force microscopy (AFM) phase and height images of dropcast films were recorded at room temperature using a Digital Instruments Multimode Nanoscope IV operating in the tapping regime mode using silicon cantilever tips (PPP-NCHR, 204–497 kHz, 10–130 N m−1). Images were processed using Gwyddion software (version 2.43). Water contact angles were measured on an OCA30 (DataPhysics) at room temperature. Water droplets (4 μL) were applied on the dropcast films and the angle of the water–air–polymer interface was measured after 30 seconds. Arnow's test was performed to detect catechol residues within the dropcast films.28 In this assay, the following components are sequentially added to produce a red/brown color in the presence of phenolic hydroxyl groups: 0.5 mL HCl (0.5 M), 0.5 mL nitrite-molybdate (0.02 v/v%), 0.5 mL NaOH (1 M), and 0.5 mL distilled water. Images were taken with a digital camera (Canon G16) and contrast was manually adapted to highlight the differences between UPy-Priplast films without and with UPy-DOPA or UPy-PCL films without and with UPy-DOPA.
5)) were dropcast on silanized glass coverslips. Dropcast films were dried in vacuum at room temperature overnight. Fibronectin-coated cover glasses were prepared by incubating them in 30 μg mL−1 fibronectin in PBS for 1–2 hours. Atomic force microscopy (AFM) phase and height images of dropcast films were recorded at room temperature using a Digital Instruments Multimode Nanoscope IV operating in the tapping regime mode using silicon cantilever tips (PPP-NCHR, 204–497 kHz, 10–130 N m−1). Images were processed using Gwyddion software (version 2.43). Water contact angles were measured on an OCA30 (DataPhysics) at room temperature. Water droplets (4 μL) were applied on the dropcast films and the angle of the water–air–polymer interface was measured after 30 seconds. Arnow's test was performed to detect catechol residues within the dropcast films.28 In this assay, the following components are sequentially added to produce a red/brown color in the presence of phenolic hydroxyl groups: 0.5 mL HCl (0.5 M), 0.5 mL nitrite-molybdate (0.02 v/v%), 0.5 mL NaOH (1 M), and 0.5 mL distilled water. Images were taken with a digital camera (Canon G16) and contrast was manually adapted to highlight the differences between UPy-Priplast films without and with UPy-DOPA or UPy-PCL films without and with UPy-DOPA.
Cell culture
L9TB cardiomyocyte progenitor cells (CMPC) were immortalized by lentiviral transduction of hTert and BMI-1 (L9TB).29 CMPCs were cultured in SP++ growth medium containing M199 (Gibco) which is bicarbonate buffered, and EGM-2 BullitKit (Lonza) in a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio, supplemented with 10% fetal bovine serum (Bovogen), 1% penicillin/streptomycin (Lonza) and 1% non-essential amino acids (Gibco) at physiological pH. CMPCs were cultured on 0.1% gelatin coated PS, passaged at 80–90% confluency and seeded at a concentration of 2.6 × 104 cells per cm2. CMPCs were cultured for 1 or 7 days on dropcast films and medium was changed every 3 days.
1 volume ratio, supplemented with 10% fetal bovine serum (Bovogen), 1% penicillin/streptomycin (Lonza) and 1% non-essential amino acids (Gibco) at physiological pH. CMPCs were cultured on 0.1% gelatin coated PS, passaged at 80–90% confluency and seeded at a concentration of 2.6 × 104 cells per cm2. CMPCs were cultured for 1 or 7 days on dropcast films and medium was changed every 3 days.
Immunofluorescent stainings
CMPCs cultured on dropcast films were first washed with PBS, fixated in 3.7% formaldehyde (Merck) for 10 minutes, washed with PBS and permeabilized with 0.5% Triton X-100 (Merck) for 10 minutes. Non-specific binding of antibodies was minimized by incubating twice in 1% horse serum (Gibco) for 10 minutes. Cells were then incubated with primary antibodies in 10% horse serum and a Net-gel buffer (50 mM Tris pH = 7.4, 150 mM NaCl, 5 mM EDTA, 0.05% NP-40, 0.25% gelatin in milliQ) overnight at 4 °C. Subsequently, cells were first washed with Net-gel and incubated with the secondary antibody for 1 hour in Net-gel followed by incubation with DAPI (0.1 μg mL−1) in PBS for 5 minutes. Finally, samples were washed and mounted on cover glasses with mowiol (Sigma). Information regarding, primary and secondary antibodies are listed in the ESI.† Samples were imaged with a confocal laser scanning microscope (Zeiss LSM510 META NLO).Viability assay and collagen staining
Viability assay was performed using Calcein-AM (Fluka) and propidium iodide (PI, Invitrogen). CMPCs cultured on dropcast films were incubated with serum-free medium with 10 μM Calcein-AM and PI for 30 minutes and imaged using a fluorescent microscope (Zeiss Axiovert 200 M). Cells were quantified using Mathematica software (Wolfram). Collagen was stained using a CNA35 probe linked to an mCherry dye (CNA35-mCherry).30 Cells were incubated with 1 μM CNA35-mCherry in standard culture medium for 30 minutes and imaged using a confocal laser scanning microscope (Leica TCS SP5X).Gene expression analysis
CMPCs were cultured for 7 days on UPy-PCL, UPy-PCL + 10 mol% UPy-DOPA and UPy-Priplast + 10 mol% UPy-DOPA and compared to CMPCs cultured on fibronectin-coated cover glass. Total RNA was isolated using the Qiagen RNAeasy isolation kit (Qiagen). cDNA was synthesized for 250 ng RNA per sample using the MMLV (Bio-Rad). Primers for quantitative PCR (qPCR) were designed using qPrimerDepot31 and BLAST. cDNA samples were subjected to qPCR using iQTM SYBR@ Green Supermix (Bio-Rad) and the Bio-Rad IQ™5 detection system (Version 1.6). Primer sequences and annealing temperatures can be found in the ESI.†Statistical analysis
Data are presented as mean ± standard deviation (SD). These data consisted of water contact angles, cellular adhesion, viability and relative gene expression. All statistical differences were determined using a non-parametric Kruskal–Wallis test with Dunn's post hoc test. Probabilities of p < 0.05 were considered as significantly different. All statistical analyses were performed using GraphPad Prism 5 Software (GraphPad Software, Inc.).Results & discussion
Synthesis of UPy-Priplast and UPy-DOPA
Hydrophobic Priplast polymers (Mn = 4945 g mol−1, Mw = 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 350 g mol−1 and Mw/Mn = 2.30) were successfully end-functionalized with UPy groups with a yield of 72% (22 g) (Scheme 1). This resulted in a polydisperse UPy-Priplast polymer with Mn = 5.0 kg mol−1, Mw = 11.6 kg mol−1 and Mw/Mn = 2.3. UPy-Priplast polymers appeared as a clear rubbery plastic.
350 g mol−1 and Mw/Mn = 2.30) were successfully end-functionalized with UPy groups with a yield of 72% (22 g) (Scheme 1). This resulted in a polydisperse UPy-Priplast polymer with Mn = 5.0 kg mol−1, Mw = 11.6 kg mol−1 and Mw/Mn = 2.3. UPy-Priplast polymers appeared as a clear rubbery plastic.
Synthesis of UPy-modified catechol functionalities (UPy-DOPA) was achieved by employing a peptide coupling strategy, which started by activating the carboxylic acid on UPy-COOH using HATU and pyridine (Scheme S2†).26 The activated carboxylic acid was then able to react to the amine on 3,4-dihydroxyphenethylamine to form the final UPy-DOPA product with a yield of 87% (166 mg). UPy-DOPA appeared as a sticky white-yellow solid.
Characterization of UPy-Priplast
The thermal behavior of Priplast polymers was studied, showing a glass transition temperature Tg of −56.3 °C, a crystallization temperature Tc of −33 °C (ΔHc is 1.6 J g−1), and a melting temperature Tm of −21.2 °C (ΔHm is 1.4 J g−1). On functionalization with UPy-moieties, the thermal behavior of UPy-Priplast clearly changes, with a glass transition temperature Tg of −51.9 °C, a crystallization temperature Tc of 37.1 °C (ΔHc is 4 J g−1), and a melting temperature Tm of 77.3 °C (ΔHm is 5.6 J g−1). This Tm is attributed to the formation of UPy-assemblies. Based on these results it can be concluded that UPy-Priplast is a semi-crystalline polymer at cell culture temperatures (37 °C). Additionally, the UPy-assemblies are present at cell culture temperatures since the Tm is 77.3 °C.The mechanical properties of UPy-Priplast elastomer films were studied. Films with thicknesses of 0.14 ± 0.02 mm had Young's moduli of 10.6 ± 1.7 MPa, a stress-at-break of 0.84 ± 0.11 MPa and a strain-at-break of 11.1 ± 1.3%. These values are slightly lower compared to the mechanical properties of previously reported UPy-PCL based films.25 However, this is expected since PCL itself crystallizes and the UPy-aggregate formation in UPy-PCL is guided by the presence of urea functionalities instead of urethanes as present in UPy-Priplast.
Formulation of catechol modified supramolecular films
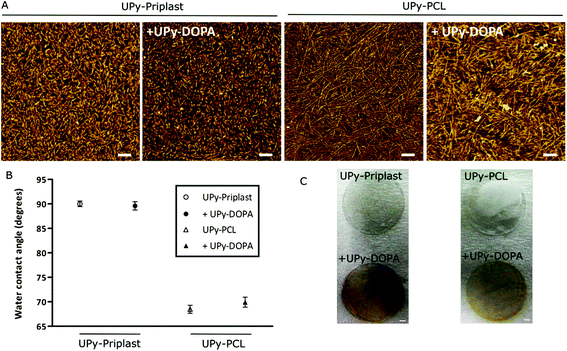
Polymer solutions were prepared in organic solvent and simple mixing of different UPy-polymers with UPy-DOPA resulted in consistent polymer mixtures. Dropcast films were then formulated by casting the polymer solution on glass surfaces and following evaporation of the solvents resulted in supramolecular films. The supramolecular structure of the dropcast films with and without UPy-DOPA additives was investigated with AFM (Fig. 1A). Short and regular fibers are observed on UPy-Priplast films at the nanometer scale compared to longer fibers on UPy-PCL films (Fig. 1A). This originates from stacking of hydrogen bonded UPy-dimers in the lateral direction which is aided by additional hydrogen bonding between the urethane (in UPy-Priplast) or urea (in UPy-PCL) functionalities (Scheme 1).32,33 The difference in fiber morphology is attributed to the presence of the urethane functionality in the UPy-Priplast material that is able to form one hydrogen bond, in contrast to the urea functionality in the UPy-PCL material which is able to form two hydrogen bonds (Scheme 1).34 Upon mixing of 10 mol% UPy-DOPA with UPy-Priplast or UPy-PCL, a similar fibrous morphology can be observed as for the pristine polymer films (Fig. 1A). This indicates successful incorporation of UPy-DOPA within the UPy-stacks of UPy-Priplast and UPy-PCL. Additionally, the AFM height images show similar surface morphology on the bioactivated UPy-polymers compared to the pristine UPy-polymer films, which also indicates successful incorporation of UPy-DOPA (Fig. S1†).Water contact angle measurements showed no difference in hydrophilicity when 10 mol% UPy-DOPA was mixed with either pristine polymers of UPy-Priplast (89.6 ± 0.8° vs. 90.0 ± 0.6°, with and without UPy-DOPA, respectively) or UPy-PCL (69.9 ± 1.0° vs. 68.5 ± 0.8°, with and without UPy-DOPA, respectively) (Fig. 1B). This is in contrast with previous reports, where a significant decrease in WCA was observed after modification of hydrophobic surfaces with catechol molecules via polydopamine.13 This could be due to higher amount of catechols in polydopamine-modified material surfaces. Dropcast films consisting of pristine UPy-DOPA showed relatively hydrophilic water contact angles of 50.3 ± 2.3°. This might indicate that the hydrophilic PEG linker and catechol groups of UPy-DOPA is initially not at the surface, and in the presence of cells and culture conditions it slowly re-assembles to the surface. To determine the presence of catechol functionalities in the dropcast films, an Arnow's test was performed (Fig. 1C). Arnow's test is a colorimetric assay which is based on the nitrosation of hydroxyl-containing benzenes which results in a color change. A clear red/brown color change was observed for dropcast films containing UPy-DOPA, which indicates the presence of oxidated catechol moieties. However, this apparently does not result in changes in hydrophilicity of the surface. In conclusion, these results show successful bioactivation of UPy-Priplast and UPy-PCL materials using UPy-DOPA additives.
CMPC adhesion and viability
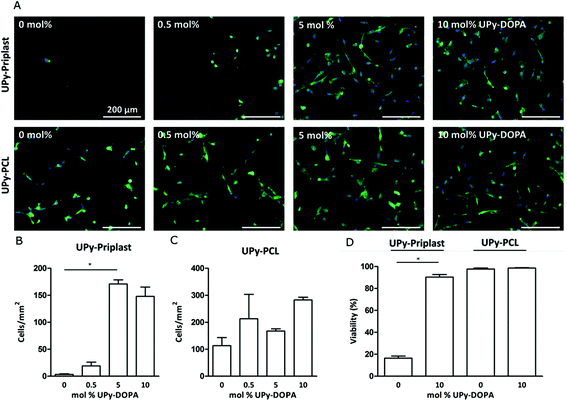
Incorporation and bioactivity of UPy-DOPA within pristine UPy-based polymers was tested at a cellular level by investigating CMPC adhesion after 24 hours of culturing (Fig. 2A). On UPy-Priplast CMPCs did not adhere after 24 hours (Fig. 2A). Upon increase in concentration of UPy-DOPA, an increase in CMPC adhesion was observed for 5 mol% and 10 mol%. This matches with the total amount of cells per mm2, which significantly increases to 170.9 ± 13.4 for 5 mol% and 147.9 ± 29.9 for 10 mol% UPy-DOPA (Fig. 2B). The increase in adhesion and cell number is either due to, (i) an increase in the number of bound serum proteins or (ii) an increase in the adsorption strength of serum proteins on UPy-Priplast. In literature, improved adhesion on catechol-functionalized surfaces is interpreted by the ability to immobilize thiol/amine containing biomolecules via Michael-type addition or Schiff-base reactions to o-quinones.18 In this study, no sodium periodate or basic solutions were used, which are required to oxidize catechols to reactive o-quinones.6 Another explanation could be that serum proteins are stabilized by the hydroxy groups of the catechol via hydrogen bonding. Furthermore, on hydrophobic surfaces, proteins tend to denature and therefore they might lose some of their adhesive properties.35 As a control group, both UPy-Priplast and UPy-PCL were functionalized with UPy-MeO, an analogue guest molecule lacking catechol functionality, to confirm the effect of catechols at the surface (Fig. S2†). On UPy-Priplast functionalized with UPy-MeO, no improvement in the adhesion of CMPCs was observed, proving the effect of catechol functionalities that improve cellular adhesion on UPy-Priplast films (Fig. S3†). On pristine UPy-PCL CMPCs showed adhesion and spreading (Fig. 2A). This is in agreement with previous studies where 3T3 fibroblasts showed adhesion and spreading when cultured on pristine UPy-PCL.25 CMPCs showed a slight increase in spreading upon increase in molar concentration of UPy-DOPA to 10 mol%. Additionally, the amount of cells per mm2 slightly increased upon increase of UPy-DOPA concentration, however not significantly (Fig. 2C). Similarly, UPy-PCL functionalized with UPy-MeO showed no improvement of cell adhesion (Fig. S4†). Next, viability of CMPC was assessed on UPy-based films after 24 hours of culturing (Fig. 2D and S5†). On UPy-Priplast CMPCs showed low viability (16.5 ± 3.1%), which is primarily due to the loss of cellular adhesion after 24 hours of culturing. Upon incorporation of UPy-DOPA CMPC adhesion significantly increased (90.4 ± 4.1%). On UPy-PCL CMPCs showed high viability and upon incorporation of UPyDOPA, viability remained unchanged (97.7 ± 1.7% and 98.6 ± 0.6%, respectively) (Fig. 2D). However, the absolute number of adhered cells did increase upon incorporation of 10 mol% UPy-DOPA (data not shown). Improvement of CMPC adhesion was achieved by modular incorporation of low amounts of UPy-DOPA (1.67 mg ml−1) in UPy-based polymers, excluding the use of oxidating agents or basic pH.Cardiac ECM gene expression
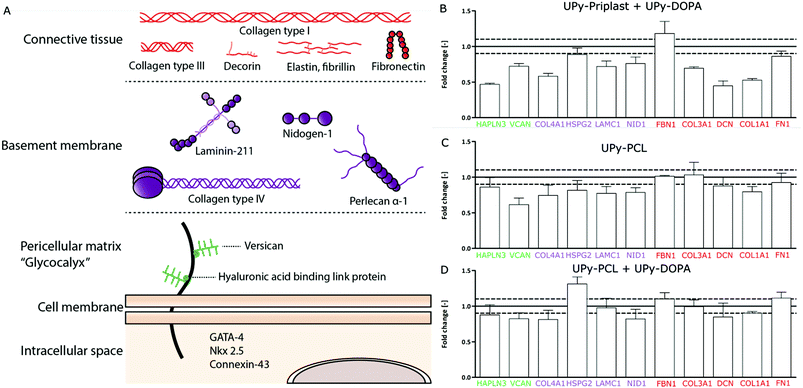
Recently, CMPCs have proven to be a potential cell source for cardiac tissue engineering approaches due to their ability to proliferate, differentiate into cardiomyocytes and their capacity to produce cardiac ECM components.29,36,37 To investigate the ECM formation capabilities of CMPCs on different supramolecular surfaces, cardiac specific ECM gene expression was studied. For simplification, the cardiac ECM was divided into three categories, (1) the pericellular matrix, (2) basement membrane and (3) connective tissue layer (Fig. 3A). The pericellular matrix consists of hyaluronan and proteoglycan link protein 3 (HAPLN3) and versican (VCAN). The basement membrane consists of collagen type IV (COL4A1), proteoglycan α-1 (HSPG2), laminin γ-1 (LAMC1) and nidogen (NID1). Finally, the connective tissue layer consists of fibrillin-1 (FBN1), collagen type III (COL3A1), decorin (DCN), collagen type I (COL1A1) and fibronectin (FN1). CMPCs cultured for 7 days on the different supramolecular surfaces were compared to CMPCs cultured on fibronectin-coated glass to check the effect of the supramolecular surfaces on gene expression levels (Fig. 3B–D). Due to high cell death on pristine UPy-Priplast surfaces after 1 day, no cells remained and gene expression was not possible to be measured for this condition after 7 days (Fig. 2D). On UPy-Priplast functionalized with UPy-DOPA, low gene expression for the ECM components HAPLN3, COL4A1, DCN and COL1A1 was seen compared to CMPCs cultured on fibronectin-coated cover glass (Fig. 3D). This might be due to the lower amount of CMPCs on UPy-Priplast based films compared to UPy-PCL based films after 7 days of culture (data not shown). Nevertheless, cardiac markers which are identical for CMPC were expressed and maintained on all surfaces (Fig. S6†). On pristine UPy-PCL, genes expressed for ECM components found in the pericellular matrix, basement membrane and connective tissue layer, showed similar levels of expression compared to CMPCs cultured on fibronectin-coated glass (Fig. 3B). Similarly, on UPy-DOPA functionalized UPy-PCL films, gene expression of all cardiac categories remained similar compared to CMPCs cultured on fibronectin-coated glass (Fig. 3C). These results show the feasibility to maintain CMPC-based cardiac ECM formation through the preservation of ECM gene expression and CMPC phenotype on catechol-functionalized supramolecular surfaces.Cardiac ECM production
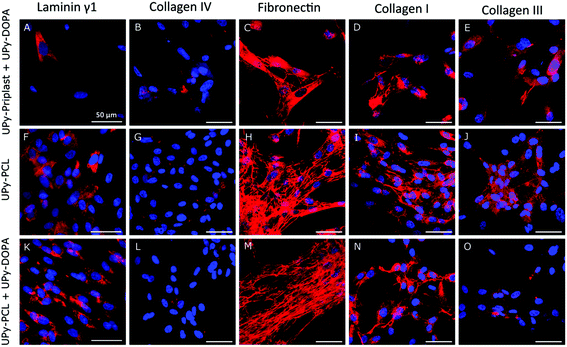
Cardiac specific protein production and localization on supramolecular surfaces and the effect of catechol incorporation was studied (Fig. 4). Similarly, due to high cell death on pristine UPy-Priplast surfaces after 1 day of culture, ECM production was not detected after 7 days (Fig. 2D). The basement membrane component laminin γ1 was located in the cytosol (Fig. 4A, F and K). Similarly, collagen IV was observed on all surfaces (Fig. 4B, G and L). The presence of extracellular fibronectin was observed on all surfaces, however no difference were observed when catechol functionalities were incorporated (Fig. 4C, H and M). Further analysis should be performed to assess fibronectin immobilization on catechol-functionalized supramolecular surfaces, since fibronectin production is a crucial ECM component which has been shown to improve matrix assembly and cardiac progenitor cell proliferation and alignment.38,39 Furthermore, extracellular collagen type I was observed on all surfaces (Fig. 4D, I and N). Additionally, the presence of extracellular collagen was observed on all surfaces, which was confirmed with CNA35-mCherry staining (Fig. S7†).40 Lastly, collagen type III was mostly observed within the cytosol of CMPCs (Fig. 4E, J and O). With these results, it can be concluded that ECM production by CMPCs is preserved on catechol-functionalized supramolecular surfaces.Conclusions
Successful formulation and development of adhesive supramolecular surfaces using UPy-DOPA was achieved. This was realized by modularly incorporating UPy-DOPA with non-cell adhesive UPy-Priplast and cell-adhesive UPy-PCL, thereby, creating improved cellular adhesive supramolecular biomaterials. Additionally, these supramolecular biomaterials showed the capability to support the preservation of endogenous ECM gene and protein expression by CMPCs, which is of high relevance for cardiac tissue engineering approaches.Acknowledgements
Marie-José Goumans is gratefully acknowledged for the L9TB CMPC. We also thank Marleen Kamperman for the useful discussions. We acknowledge the support from the Netherlands Cardiovascular Research Initiative (CVON 2012-01). The Dutch Heart Foundation, Dutch Federation of University Medical Centers, the Netherlands Organization for Health Research and Development and the Royal Netherlands Academy of Sciences. Additionally, this work was financially supported by the European Research Council (FP7/2007–2013) ERC Grant Agreement 308045, the Ministry of Education, Culture and Science (Gravity program 024.001.03), and ZonMW as part of the LSH 2Treat program (project number 436001003).Notes and references
- B. P. Lee, P. B. Messersmith, J. N. Israelachvili and J. H. Waite, Annu. Rev. Mater. Res., 2011, 41, 99–132 CrossRef CAS PubMed.
- H. Lee, B. P. Lee and P. B. Messersmith, Nature, 2007, 448, 338–341 CrossRef CAS PubMed.
- M. D. Bartlett, A. B. Croll and A. J. Crosby, Adv. Funct. Mater., 2012, 22, 4985–4992 CrossRef CAS.
- H. Lee, S. M. Dellatore, W. M. Miller and P. B. Messersmith, Science, 2007, 318, 426–430 CrossRef CAS PubMed.
- R. J. Stewart, T. C. Ransom and V. Hlady, J. Polym. Sci., Part B: Polym. Phys., 2011, 49, 757–771 CrossRef CAS PubMed.
- J. Yang, M. A. Cohen Stuart and M. Kamperman, Chem. Soc. Rev., 2014, 43, 8271–8298 RSC.
- Q. Lin, D. Gourdon, C. J. Sun, N. Holten-Andersen, T. H. Anderson, J. H. Waite and J. N. Israelachvili, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 3782–3786 CrossRef CAS PubMed.
- N. K. Kaushik, N. Kaushik, S. Pardeshi, J. G. Sharma, S. H. Lee and E. H. Choi, Mar. Drugs, 2015, 13, 6792–6817 CrossRef CAS PubMed.
- L. Li, W. Smitthipong and H. Zeng, Polym. Chem., 2015, 6, 353–358 RSC.
- C. E. Brubaker and P. B. Messersmith, Langmuir, 2012, 28, 2200–2205 CrossRef CAS PubMed.
- S. K. Madhurakkat Perikamana, J. Lee, Y. Bin Lee, Y. M. Shin, E. J. Lee, A. G. Mikos and H. Shin, Biomacromolecules, 2015, 16, 2541–2555 CrossRef CAS PubMed.
- Y. M. Shin, I. Jun, Y. M. Lim, T. Rhim and H. Shin, Macromol. Mater. Eng., 2013, 298, 555–564 CrossRef CAS.
- S. H. Ku, J. Ryu, S. K. Hong, H. Lee and C. B. Park, Biomaterials, 2010, 31, 2535–2541 CrossRef CAS PubMed.
- N. Li, G. Chen, J. Liu, Y. Xia, H. Chen, H. Tang, F. Zhang and N. Gu, ACS Appl. Mater. Interfaces, 2014, 6, 17134–17143 CAS.
- C. E. Brubaker and P. B. Messersmith, Biomacromolecules, 2011, 12, 4326–4334 CrossRef CAS PubMed.
- C. Lee, J. Shin, J. S. Lee, E. Byun, J. H. Ryu, S. H. Um, D. I. Kim, H. Lee and S. W. Cho, Biomacromolecules, 2013, 14, 2004–2013 CrossRef CAS PubMed.
- H. S. Kim, H. O. Ham, Y. J. Son, P. B. Messersmith and H. S. Yoo, J. Mater. Chem. B, 2013, 1, 3940–3949 RSC.
- J. S. Choi, P. B. Messersmith and H. S. Yoo, Macromol. Biosci., 2014, 14, 270–279 CrossRef CAS PubMed.
- P. Y. W. Dankers, J. M. Boomker, A. Huizinga-van der Vlag, E. Wisse, W. P. J. Appel, F. M. M. Smedts, M. C. Harmsen, A. W. Bosman, W. Meijer and M. J. A. van Luyn, Biomaterials, 2011, 32, 723–733 CrossRef CAS PubMed.
- B. B. Mollet, M. Comellas-Aragonès, A. J. H. Spiering, S. H. M. Söntjens, E. W. Meijer and P. Y. W. Dankers, J. Mater. Chem. B, 2014, 2, 2483 RSC.
- G. C. Van Almen, H. Talacua, B. D. Ippel, B. B. Mollet, M. Ramaekers, M. Simonet, A. I. P. M. Smits, C. V. C. Bouten, J. Kluin and P. Y. W. Dankers, Macromol. Biosci., 2016, 16, 350–362 CrossRef CAS PubMed.
- A. C. H. Pape, B. D. Ippel and P. Y. W. Dankers, Langmuir, 2017, 33(16), 4076–4082 CrossRef CAS PubMed.
- P. Goichberg, J. Chang, R. Liao and A. Leri, Antioxid. Redox Signaling, 2014, 21, 2002–2017 CrossRef CAS PubMed.
- S. Koudstaal, S. J. Jansen of Lorkeers, R. Gaetani, J. M. I. H. Gho, F. J. van Slochteren, J. P. G. Sluijter, P. A. Doevendans, G. M. Ellison and S. A. J. Chamuleau, Stem Cells Transl. Med., 2013, 2, 434–443 CrossRef PubMed.
- P. Y. W. Dankers, M. C. Harmsen, L. a. Brouwer, M. J. A. Van Luyn and E. W. Meijer, Nat. Mater., 2005, 4, 568–574 CrossRef CAS PubMed.
- I. De Feijter, O. J. G. M. Goor, S. I. S. Hendrikse, M. Comellas-Aragonès, S. H. M. Söntjens, S. Zaccaria, P. P. K. H. Fransen, J. W. Peeters, L. G. Milroy and P. Y. W. Dankers, Synlett, 2015, 2707–2713 CAS.
- H. M. Keizer, R. Van Kessel, R. P. Sijbesma and E. W. Meijer, Polymer, 2003, 44, 5505–5511 CrossRef CAS.
- L. E. Arnow, J. Comp. Educ., 1937, 118, 531–537 CAS.
- A. M. Smits, P. van Vliet, C. H. Metz, T. Korfage, J. P. G. Sluijter, P. A. Doevendans and M.-J. Goumans, Nat. Protoc., 2009, 4, 232–243 CrossRef CAS PubMed.
- S. J. A. Aper, A. C. C. Van Spreeuwel, M. C. Van Turnhout, A. J. Van Der Linden, P. A. Pieters, N. L. L. Van Der Zon, S. L. De La Rambelje, C. V. C. Bouten and M. Merkx, PLoS One, 2014, 9, 1–21 Search PubMed.
- W. Cui, D. D. Taub and K. Gardner, Nucleic Acids Res., 2007, 35, D805–D809 CrossRef CAS PubMed.
- W. P. J. Appel, G. Portale, E. Wisse, P. Y. W. Dankers and E. W. Meijer, Macromolecules, 2011, 44, 6776–6784 CrossRef CAS.
- E. Wisse, A. J. H. Spiering, P. Y. W. Dankers, B. Mezari, P. C. M. M. Magusin and E. W. Meijer, J. Polym. Sci., Part A: Polym. Chem., 2011, 49, 1764–1771 CrossRef CAS.
- H. Kautz, D. J. M. Van Beek, R. P. Sijbesma and E. W. Meijer, Macromolecules, 2006, 39, 4265–4267 CrossRef CAS.
- J. li Zhai, L. Day, M. I. Aguilar and T. J. Wooster, Curr. Opin. Colloid Interface Sci., 2013, 18, 257–271 CrossRef.
- N. A. M. Bax, M. H. van Marion, B. Shah, M.-J. Goumans, C. V. C. Bouten and D. W. J. van der Schaft, J. Mol. Cell. Cardiol., 2012, 53, 497–508 CrossRef CAS PubMed.
- M. H. van Marion, N. A. M. Bax, M. C. van Turnhout, A. Mauretti, D. W. J. van der Schaft, M. J. T. H. Goumans and C. V. C. Bouten, J. Mol. Cell. Cardiol., 2015, 87, 79–91 CrossRef CAS PubMed.
- A. Hielscher, K. Ellis, C. Qiu, J. Porterfield and S. Gerecht, PLoS One, 2016, 11, e0147600 Search PubMed.
- K. M. French, J. T. Maxwell, S. Bhutani, S. Ghosh-Choudhary, M. J. Fierro, T. D. Johnson, K. L. Christman, W. R. Taylor and M. E. Davis, Stem Cells Int., 2016, 2016, 1–11 CrossRef PubMed.
- J. Glowacki and S. Mizuno, Biopolymers, 2008, 89, 338–344 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7bm00407a |
| This journal is © The Royal Society of Chemistry 2017 |