Personalized protein corona on nanoparticles and its clinical implications
Claudia
Corbo
 a,
Roberto
Molinaro
b,
Mateen
Tabatabaei
a,
Omid C.
Farokhzad
*ac and
Morteza
Mahmoudi
a,
Roberto
Molinaro
b,
Mateen
Tabatabaei
a,
Omid C.
Farokhzad
*ac and
Morteza
Mahmoudi
 *ad
*ad
aCenter for Nanomedicine and Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA. E-mail: ofarokhzad@bwh.harvard.edu; mmahmoudi@bwh.harvard.edu
bCenter for Biomimetic Medicine, Houston Methodist Research Institute, Houston, TX, USA
cKing Abdulaziz University, Jeddah 21589, Saudi Arabia
dNanotechnology Research Center, Faculty of Pharmacy, Tehran University of Medical Sciences, Tehran, Iran
First published on 30th January 2017
Abstract
It is now well understood that once in contact with biological fluids, nanoscale objects lose their original identity and acquire a new biological character, referred to as a protein corona. The protein corona changes many of the physicochemical properties of nanoparticles, including size, surface charge, and aggregation state. These changes, in turn, affect the biological fate of nanoparticles, including their pharmacokinetics, biodistribution, and therapeutic efficacy. It is progressively being accepted that even slight variations in the composition of a protein source (e.g., plasma and serum) can substantially change the composition of the corona formed on the surface of the exact same nanoparticles. Recently it has been shown that the protein corona is strongly affected by the patient's specific disease. Therefore, the same nanomaterial incubated with plasma proteins of patients with different pathologies adsorb protein coronas with different compositions, giving rise to the concept of personalized protein corona. Herein, we review this concept along with recent advances on the topic, with a particular focus on clinical relevance.
Introduction
In a physiological milieu, a shell of active biomolecules forms around NPs referred to as a protein corona (PC).1 The PC, also known as the biomolecular corona, is mainly composed of proteins. However, researchers also expect to find other biomolecules such as lipids, metabolites, and sugars in future investigations.2,3 The phenomenon of PC formation is frequent among all known NPs, except for a few with coatings such as zwitterionic,4,5 and has an important impact on the NP's properties. PC-coated NPs strongly differ in size, surface charge and aggregation state compared to pristine NPs.6–8 PC composition is affected by both NP characteristics (e.g., physicochemical properties) and environmental factors (e.g., temperature, protein source, type of disease present). The effect of NPs’ physicochemical properties on corona formation have been extensively examined and already comprehensively reviewed by us and others.1,6,9–13 However, many environmental factors (e.g., protein and ionic concentrations,14 temperature,15 origin [human or murine],16 protein source [fetal bovine serum, serum or plasma],17 choice of anticoagulant,16 and flow status [dynamic vs. static]18) have been only recently introduced19 while several others (e.g., personalized protein corona and disease-specific protein corona)20 are not yet understood. It is well known that the dynamic nature of the vascular system and its constituents are altered in pathological conditions.21 Importantly, changes in the abundance of several proteins in the plasma have been used as biomarkers to help diagnose and/or forecast the severity of different diseases.22–25 Keeping in mind that each disease is characterized by different plasma proteomes, our group has hypothesized and demonstrated that different diseases induce the formation of different PCs on the same nanomaterial.20 We investigated the effects of alterations in the plasma proteome due to different pathological conditions on the PC composition of commercially available NPs, thus introducing the novel concept of “personalized protein corona” (PPC) (Fig. 1).20 Other groups have proven that the plasma proteome is not always consistent, even in healthy individuals, either in terms of composition or abundance of certain proteins.26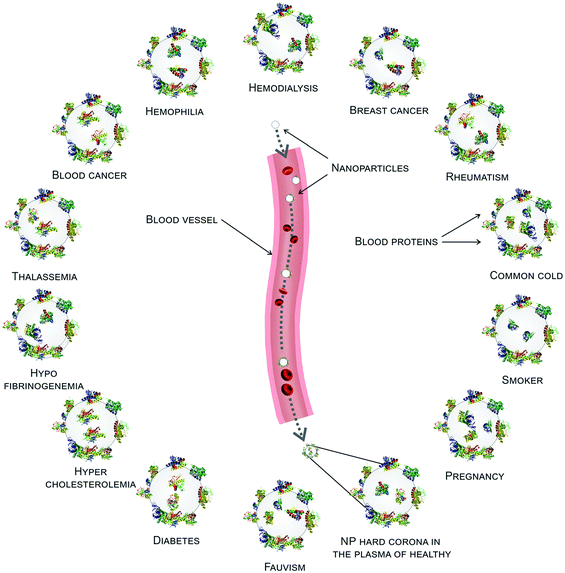 | ||
| Fig. 1 Disease-dependent personalized protein corona. The biological environment that comes into contact with NPs affects the protein corona (PC): plasma protein alterations due to disease conditions affect the type, amount, and conformation of proteins that compose the corona. Reproduced from ref. 20 with permission from Royal Society of Chemistry. | ||
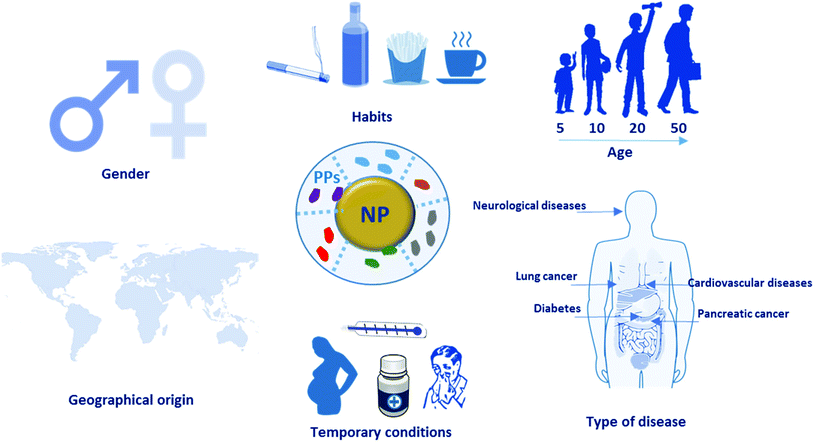
Herein, we expand the concept of “personalized protein corona” to include not only the PC formed around NPs during incubation with plasma from human patients with specific diseases, but also the PC formed from plasma of healthy individuals of different ages and genders or with certain lifestyles, habits, temporary conditions, and geographical origins (Fig. 2). Despite having been thus far overlooked, the PPC plays a crucial role in the biological fate of nanoparticles, and represents one possible explanation for the unsuccessful clinical results of some nano-products in clinical trials. In this mini-review, we highlight the importance of this new concept for avoiding misinterpretation of PC data, report recent studies on the PPC and discuss its impact in the clinical setting.
Plasma proteins as disease biomarkers
Human blood can contain biomarkers for various diseases.27–32 In the case of cancer, blood components such as some specific cell types,33 peptides,34 microRNAs,35 metabolites36 and proteins37 are used as indicators of pathological status.27 Plasma protein profiles have been widely investigated as biomarkers. In certain cases, even the level of post-translational modifications of plasma proteins has been used to distinguish patients with a given disease from healthy individuals.38,39 For example, highly altered glycosylation levels (i.e., sialylation and fucosylation) of complement C3, histidine-rich glycoprotein and kininogen-1 have been correlated with colorectal cancer progression.38 On the other hand, carbonylation of plasma proteins (e.g., VEGFR-2, MMP-1, and complement C5) is associated with obesity-induced type 2 diabetes mellitus.39Some plasma proteins are components of the acute/chronic inflammatory response. These “inflammation-sensitive plasma proteins” (ISPs) have been associated with myocardial infarction, stroke, diabetes, and prediabetes conditions (i.e., impaired glucose tolerance).40–47 In particular, increased levels of fibrinogen-alpha, α-antitrypsin, and C-reactive protein have been connected with diabetes,47 while C-reactive protein, tumor necrosis factor alpha, and other inflammatory markers have been identified as biomarkers for the prediction of cardiovascular diseases.45 Moderate systemic inflammation is associated with the pathology of chronic heart failure and causes several further problems such as myocardial remodeling and cardiac arrhythmia.48 Many epidemiological studies have connected higher plasma levels of inflammatory markers with the incidence of cardiovascular diseases.49,50 Indeed, population-based cohort studies with follow-up ∼20 years revealed that plasma levels of ISPs, including fibrinogen and alpha-1-antitrypsin, are correlated to incidence of hospitalization for heart failure.43
Another plasma protein strongly affected by the presence of inflammation is albumin, the major protein in plasma51 and, for this reason, one of the first proteins to be absorbed in the PC of the majority of known nanomaterials.52,53 Albumin is synthesized by the liver and is fundamental for the transport of hormones and vitamins throughout the body.51 Complications inducing liver malfunction and disorders change the level of albumin in the plasma. Moreover, albumin levels are also indicators of different pathological conditions, such as poor nutrition and infection.54
To further complicate the situation, several studies have also reported modulations of the levels of plasma proteins in patients with neurological disturbances.24,55–58 Immunoglobulin IgM, complement C3c, complement C4, and α-antitrypsin have all been found to be elevated in patients with depression, while less albumin and transferrin were found than in the plasma of healthy volunteers.57 Meanwhile, plasma levels of complement factor-H and alpha-2-macroglobulin in Alzheimer's disease patients are dramatically higher than in healthy people and their abundance was also correlated with the severity of the disease.56,58,59 Though these findings are certainly important for the diagnosis and prediction of all the above-mentioned diseases, they are also crucial evidence that human plasma, like other biological fluids, cannot be considered homogeneous. This concept should be incorporated into the design of PC-based studies. For instance, in planning investigations of the effect of PC formation on NPs’ targeting, distribution, and/or toxicity, it is essential to utilize plasma or sera from patients for whom there is data on the type of disease, stage, and their gender/age/lifestyle and geographical origin. These details should be reported to better interpret findings and to allow other researchers to customize their particles in a patient-specific manner.
Disease-induced variability in the protein corona composition
As mentioned above, various diseases can cause variations in the composition of the plasma proteome and/or the conformation of proteins. This, in turn, implies potential changes in the identity and conformation of the PC's components. Below we will discuss how changes in protein conformations affect the PC. Herein we introduce two recent studies featuring extensive analysis of changes in the PC composition in a wide range of diseases.20,26In the first study, PPCs formed on silica-and-polystyrene NPs were investigated using plasma derived from patients with diseases such as cancer, diabetes, hypercholesterolemia, rheumatism, fauvism, hemophilia, hypofibrinogenemia, and healthy individuals with different lifestyle factors including smoking and fat-rich diets or temporary conditions such as pregnancy.20 Protein patterns of the PCs on silver-stained SDS-PAGE gels clearly showed that the PCs associated with various diseases differed both in terms of composition and amount. Furthermore, PCs generated using plasma from patients suffering from the same disease and with the same lifestyle were quite similar, with only slight differences. Lastly, even in the case of plasma from healthy subjects of the same gender and age, PCs were not always consistent.20
The results of the first study were confirmed by Colapicchioni et al.26 (Fig. 3) in the course of evaluating PPCs formed on clinically approved liposomes (AmBisome) in gastric, breast, and pancreatic cancer patients (10 individuals per disease). The authors found that changes in the liposome's size attributable to the PC were similar between healthy volunteers and cancer patients. PC-coated liposomes were less negatively charged in pancreatic cancer patients than in breast and gastric patients, thus indicating higher levels of cationic plasma proteins. PC profiles analyzed by SDS-PAGE were similar in various cases, with specific bands more abundant in the PC associated with pancreatic cancer, confirming the surface charge findings. Among the most abundant bands, one at MW ≈ 37 kDa associated with heavy chains of immunoglobulin alpha (IgA) and immunoglobulin gamma (IgG) was identified.26 Cancer patients show important changes in the concentration of Igs, that are correlated with the presence of autoantibodies produced as immunodefense against tumors. This finding is of particular interest because it opens the way for the use of NP-PC-based technology in screens for early diagnostic tumor biomarkers (e.g. autoantibodies).60–63
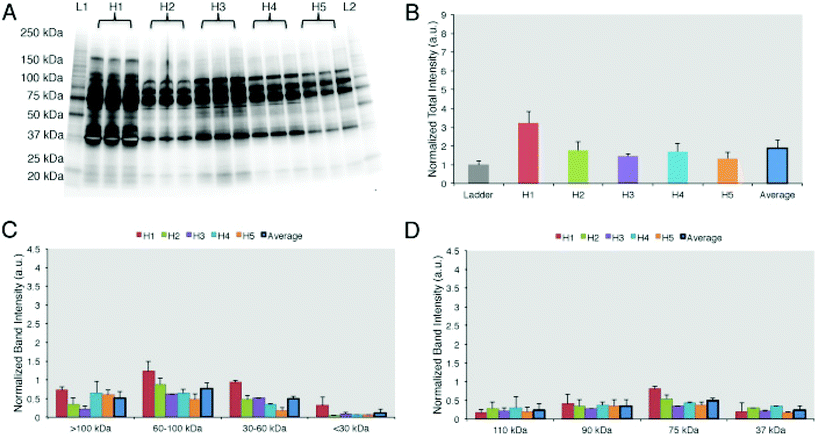 | ||
| Fig. 3 Personalized protein corona in healthy volunteers. (A) 1D-SDS PAGE image of the protein corona profiles formed around liposomes when incubated for 1 h with human plasma from five healthy volunteers. (B) Intensity of lanes analyzed by ImageJ showing that the protein coronas are different in abundance among the 5 healthy donors. (C) Band intensity according to the molecular weights. (D) The top 4 most abundant band intensities are compared among the 5 samples. Results are presented as average ± SD. Reproduced from ref. 26 with permission from Elsevier publishing group. | ||
Cancer biomarkers are found at low concentrations during the early stages of a disease, making them very difficult to detect. However, adsorption in the PC creates a “nano-concentrator”26 of proteins not easily detectable under other conditions. It is accepted that the most abundant plasma proteins are the first to be absorbed by NPs; they form a short-term layer of low-affinity proteins defined as the soft corona. However, PC formation is dynamic, and the soft corona is soon replaced by a hard corona of proteins with higher affinity to the selected NP despite being less abundant in the plasma.64 This makes these relatively rare proteins much more easily detectable as biomarkers. For example, Zheng et al.61 developed a simple PC-based assay for the early screening of prostate cancer using gold nanoparticles, which has been shown to be more specific than the current standard test for detection of early-stage prostate cancer (i.e., prostate-specific antigen test).
Disease-induced variability in protein corona conformation
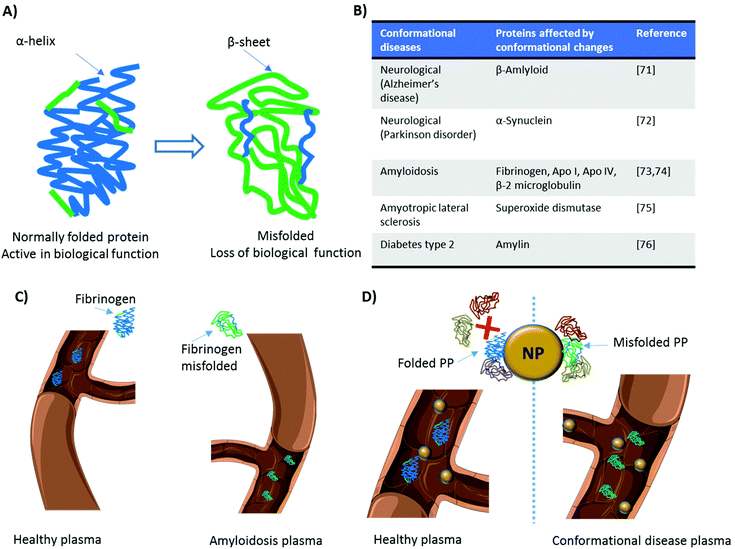
When proteins interact with NPs, changes in their conformational state can occur.65–67 The denaturation of proteins adsorbed on NPs’ surface induces the exposure of epitopes normally hidden.68 This can increase NPs’ immunogenicity and/or trigger inflammatory responses, as in the case of gold NPs functionalized with poly(acrylic) acid.69 Deng et al.69 demonstrated that functionalized gold NPs induced the unfolding of adsorbed fibrinogen. Fibrinogen possesses a region with a binding affinity toward the leukocyte receptor MAC-1. However, when it is folded, functional active fibrinogen does not interact with MAC-1 because the hydrophobic MAC-1 affinity region is facing the internal hydrophobic core.70 Instead, when bound to gold NPs, fibrinogen changed its conformation and, once unfolded, could interact with MAC-1. This interaction activated an inflammatory cascade69 and established the basis for the introduction of a novel mechanism underlying the inflammatory response to NPs, mediated by the PC.On the other hand, several pathologies arise from the presence of proteins in conformational states different from their original one, i.e., misfolded (Fig. 4A). These diseases, also known as proteinopathies or conformational diseases, include neurodegenerative disorders,71,72 amyloidosis,73,74 and many others75,76 (several examples are reported in Fig. 4B) and are generally characterized by misfolded proteins whose conformational change triggers their tendency to aggregate with each other. While several diseases can bring variations in the protein composition of plasma, conformational diseases induce changes in the conformation of proteins, thus affecting their interactions with NPs and, consequently, their PC. In theory, the plasma of a patient with a conformational disease changes not in terms of protein composition (Fig. 4C). The interaction of the same protein in two different conformations with a given NP is not the same (Fig. 4D). For this reason, it is crucial to study not only the identity but also the conformational state of proteins in the corona. As the protein corona is a complex system, current approaches (e.g., circular dichroism) to defining protein conformation cannot be employed. To overcome this shortcoming, we used fluorescence resonance energy transfer (FRET)-labeled fibronectin, enabling us to monitor the conformation of this protein in the protein corona in situ. The investigation of the molecular motifs (potentially recognized by receptors of cells) exposed by the proteins in the corona is now becoming a topic of great interest in the field.77
Disease-induced nanotoxicity
Identical NPs employed to treat patients with different diseases are characterized by different (i.e., personalized) PCs. These PPCs affect NPs’ biological fate in ways we still find hard to predict. To improve the efficiency and safety of personalized nanomaterials for clinical applications, it is crucial to elucidate how the PPCs influence the therapeutic impact of NPs. We have evaluated the therapeutic/toxic impact of graphene oxide (GO) sheets coated by PPCs obtained using plasma from human subjects with various pathological conditions.78 GO sheets have attracted the attention of many researchers due to their peculiar physicochemical properties. They are already employed in several applications, including neural network regeneration, cancer therapy, and stem cell tissue regeneration.79 GO sheets were incubated with the plasma of patients with seven different diseases: thalassemia (major and minor), blood cancer, diabetes, fauvism, rheumatism, and hypercholesterolemia. The plasma from pregnant women in the same gestation period and of similar age was also used to create a PPC on GO sheets. Then, interactions between PPC-coated GO sheets and breast cancer cell lines (MCF-7 and MDA-MB-231) were evaluated in terms of cellular toxicity, apoptosis/necrosis (Fig. 5A–J), cellular uptake, production of reactive oxygen species, cellular inflammation, and lipid peroxidation.78 The results showed significantly different effects of the PPCs-coated NPs on each of these biological processes, underlining once again that detailed information on patients’ clinical history, lifestyle, and habits must be carefully considered in the development of ad hoc-designed nanomaterials.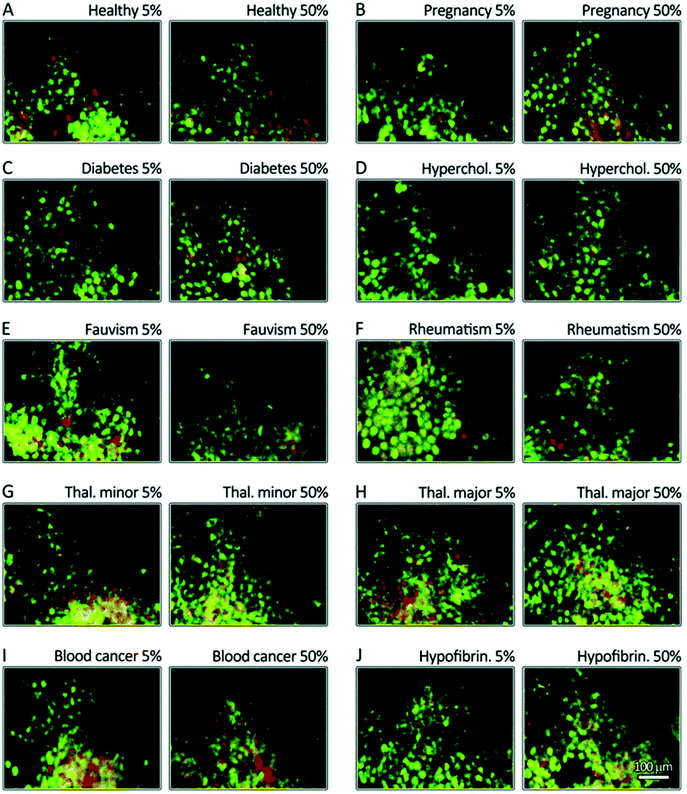 | ||
| Fig. 5 Disease-dependent apoptosis and necrosis. MCF-7 breast cancer cells were stained using propidium iodide (nuclei, green) and annexinV (membrane, yellow to red). The cells were then incubated for 24 h with corona-coated GO from patients with different diseases (A–J). The results show that the protein coronas from different diseases have different impacts on apoptosis (green) and late apoptosis/necrosis (yellow/green-red) levels. GO: graphene oxide. Reproduced from ref. 77 with permission from Royal Society of Chemistry. | ||
Similar conclusions were obtained by Shannahan and coworkers.80 The authors aimed at developing in vitro approaches to study NPs’ toxicity for cardiovascular applications. As already mentioned, individuals with cardiovascular pathologies have an altered plasma composition, potentially affecting PC formation. Moreover, many NPs are conceived for intravenous injection and are directly exposed to the vasculature. This can cause vascular inflammation, the formation of atherosclerotic plaques, and oxidative stress,81–84 demonstrating the need for a better understanding of the cardiovascular toxicity induced by NPs. PCs were obtained by incubating magnetite iron oxide NPs (Fe3O4) with normal/healthy and lipid-rich rat serum.80 Authors first tested whether the lipid-rich serum induced the formation of a different PC (PPC) compared with the normal serum, then investigated the toxicological consequences induced by PPCs. Rat aortic endothelial cells (RAECs) were incubated with PC-coated NPs under flow conditions to better mimic the in vivo scenario. PCs from both sera induced a slight increase in the hydrodynamic diameters of NPs. As expected, the amount of cholesterol in the PC formed using the lipid-rich serum was higher than that of the PC formed from normal serum. The two PCs had 92 protein components in common, while 29 unique proteins were identified only in the hyperlipidemic PC. Both normal and lipid-rich PCs reduced the association of NPs with RAECs, while the PC formed using lipid-rich serum induced intensified endothelial cell activation and inflammatory response (i.e. increased expression of IL-6, tumor necrosis factor alpha, VCAM-1),80 demonstrating the importance of physiological impact in determining NP biological response.
Clinical relevance of personalized protein corona
Despite recent progress in nanomedicine, only a few NPs have been assessed in clinical trials, and even fewer have reached clinical practice. Recent findings show that one of the reasons behind the incongruence between experimental discoveries and clinical outcomes lies in the lack of clear information on what happens at the nano–bio interface. The formation of the PC around NPs led many research groups to consider the biological identity of NPs as a fundamental actor in the definition of NPs’ biological fate. Many studies have investigated the proteins that compose the PCs of many different organic and inorganic NPs; however, results from different laboratories sometimes did not match, even when the same nanomaterial was employed.85,86 With the introduction of the new concept of PPC, the widely accepted view that identical nanomaterials have similar or identical toxic effects on individuals with different pathological conditions should be considered outdated. Data from a single patient cannot be generalized to other patients. Additionally, each individual exhibits a distinct plasma proteome on the basis of his/her specific health conditions, gender, lifestyle, and genetic background, leading to the formation of a PPC around therapeutic NPs. Characterizing disease-related PPCs can help in the prediction of the biological outcomes of NPs and in turn speed up clinical translation. In the near future, we can expect to witness the development of disease-specific custom NPs.Conclusions
To date, the majority of investigations on phenomena at the nano–bio interface have overlooked the contribution of the physiological environment to PC formation, focused instead on the effects of the NPs’ properties on the PC. Differences in the PC based on the biological environment can help explain various discordant findings of recent years, such as (i) the considerable variability in results among different research groups analyzing PCs formed on the same NPs; (ii) variations between in vitro and in vivo studies; (iii) differences between cell lines with different media; and more importantly (iv) the unsuccessful clinical results of NPs that had shown promising in vitro and in vivo results. A large body of work must be carried out to gain a better understanding of nano–bio interactions. We believe that the novel concept of the personalized protein corona, combined with other recent findings regarding the role of the biological milieu in the PC's formation, will allow us to better design future experiments in this field and accelerate clinical translation.Competing interests statement
O. C. F. declares financial interests in Selecta Biosciences, Tarveda Therapeutics and Placon Therapeutics.Acknowledgements
This work was supported by the grants US National Institutes of Health (NIH) CA151884 (O. C. F.) and EB015419 (O. C. F.). We thank Servier Medical Art for the high-quality image bank.References
- M. P. Monopoli, C. Åberg, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2012, 7, 779–786 CrossRef CAS PubMed.
- S. Wan, P. M. Kelly, E. Mahon, H. Stöckmann, P. M. Rudd, F. Caruso, K. A. Dawson, Y. Yan and M. P. Monopoli, ACS Nano, 2015, 9, 2157–2166 CrossRef CAS PubMed.
- E. Hellstrand, I. Lynch, A. Andersson, T. Drakenberg, B. Dahlbäck, K. A. Dawson, S. Linse and T. Cedervall, FEBS J., 2009, 276, 3372–3381 CrossRef CAS PubMed.
- M. Mahmoudi, N. Bertrand, H. Zope and O. C. Farokhzad, Nano Today, 2016, 11(6), 817–832 CrossRef CAS.
- R. Safavi-Sohi, S. Maghari, M. Raoufi, S. A. Jalali, M. J. Hajipour, A. Ghassempour and M. Mahmoudi, ACS Appl. Mater. Interfaces, 2016, 8, 22808–22818 CAS.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch, F. Baldelli Bombelli and K. A. Dawson, J. Am. Chem. Soc., 2011, 133, 2525–2534 CrossRef CAS PubMed.
- S. Tenzer, D. Docter, J. Kuharev, A. Musyanovych, V. Fetz, R. Hecht, F. Schlenk, D. Fischer, K. Kiouptsi, C. Reinhardt, K. Landfester, H. Schild, M. Maskos, S. K. Knauer and R. H. Stauber, Nat. Nanotechnol., 2013, 8, 772–781 CrossRef CAS PubMed.
- W. L. Koh, P. H. Tham, H. Yu, H. L. Leo and J. C. Yong Kah, Nanomed., 2016, 11, 2275–2287 CrossRef CAS PubMed.
- C. Corbo, R. Molinaro, F. Taraballi, N. Toledano Furman, M. Sherman, A. Parodi, F. Salvatore and E. Tasciotti, Int. J. Nanomed., 2016, 11, 3049–3063 CrossRef CAS PubMed.
- C. Corbo, R. Molinaro, A. Parodi, N. E. Toledano Furman, F. Salvatore and E. Tasciotti, Nanomed., 2016, 11, 81–100 CrossRef CAS PubMed.
- M. Mahmoudi, I. Lynch, M. R. Ejtehadi, M. P. Monopoli, F. B. Bombelli and S. Laurent, Chem. Rev., 2011, 111, 5610–5637 CrossRef CAS PubMed.
- K. Saha, M. Rahimi, M. Yazdani, S. T. Kim, D. F. Moyano, S. Hou, R. Das, R. Mout, F. Rezaee, M. Mahmoudi and V. M. Rotello, ACS Nano, 2016, 10, 4421–4430 CrossRef CAS PubMed.
- A. Bigdeli, S. Palchetti, D. Pozzi, M. R. Hormozi-Nezhad, F. Baldelli Bombelli, G. Caracciolo and M. Mahmoudi, ACS Nano, 2016, 10, 3723–3737 CrossRef CAS PubMed.
- G. Caracciolo, D. Pozzi, A. L. Capriotti, C. Cavaliere, P. Foglia, H. Amenitsch and A. Laganà, Langmuir, 2011, 27, 15048–15053 CrossRef CAS PubMed.
- M. Mahmoudi, A. M. Abdelmonem, S. Behzadi, J. H. Clement, S. Dutz, M. R. Ejtehadi, R. Hartmann, K. Kantner, U. Linne, P. Maffre, S. Metzler, M. K. Moghadam, C. Pfeiffer, M. Rezaei, P. Ruiz-Lozano, V. Serpooshan, M. A. Shokrgozar, G. U. Nienhaus and W. J. Parak, ACS Nano, 2013, 7, 6555–6562 CrossRef CAS PubMed.
- S. Schöttler, K. Klein, K. Landfester and V. Mailänder, Nanoscale, 2016, 8, 5526–5536 RSC.
- D. Pozzi, G. Caracciolo, A. L. Capriotti, C. Cavaliere, G. La Barbera, T. J. Anchordoquy and A. Laganà, J. Proteomics, 2015, 119, 209–217 CrossRef CAS PubMed.
- D. Pozzi, G. Caracciolo, L. Digiacomo, V. Colapicchioni, S. Palchetti, A. L. Capriotti, C. Cavaliere, R. Zenezini Chiozzi, A. Puglisi and A. Laganà, Nanoscale, 2015, 7, 13958–13966 RSC.
- M. Mahmoudi, S. E. Lohse, C. J. Murphy, A. Fathizadeh, A. Montazeri and K. S. Suslick, Nano Lett., 2014, 14, 6–12 CrossRef CAS PubMed.
- M. J. Hajipour, S. Laurent, A. Aghaie, F. Rezaee and M. Mahmoudi, Biomater. Sci., 2014, 2, 1210 RSC.
- P. Rajendran, T. Rengarajan, J. Thangavel, Y. Nishigaki, D. Sakthisekaran, G. Sethi and I. Nishigaki, Int. J. Biol. Sci., 2013, 9, 1057–1069 CrossRef CAS PubMed.
- H. C. Beck, M. Overgaard and L. Melholt Rasmussen, Transl. Proteomics, 2015, 7, 40–48 CrossRef CAS.
- S. Surinova, R. Schiess, R. Hüttenhain, F. Cerciello, B. Wollscheid and R. Aebersold, J. Proteome Res., 2011, 10, 5–16 CrossRef CAS PubMed.
- S. J. Kiddle, C. J. Steves, M. Mehta, A. Simmons, X. Xu, S. Newhouse, M. Sattlecker, N. J. Ashton, C. Bazenet, R. Killick, J. Adnan, E. Westman, S. Nelson, H. Soininen, I. Kloszewska, P. Mecocci, M. Tsolaki, B. Vellas, C. Curtis, G. Breen, S. C. R. Williams, S. Lovestone, T. D. Spector and R. J. B. Dobson, Transl. Psychiatry, 2015, 5, e584 CrossRef CAS PubMed.
- G. Schley, C. Köberle, E. Manuilova, S. Rutz, C. Forster, M. Weyand, I. Formentini, R. Kientsch-Engel, K.-U. Eckardt and C. Willam, PLoS One, 2015, 10, e0145042 Search PubMed.
- V. Colapicchioni, M. Tilio, L. Digiacomo, V. Gambini, S. Palchetti, C. Marchini, D. Pozzi, S. Occhipinti, A. Amici and G. Caracciolo, Int. J. Biochem. Cell Biol., 2016, 75, 180–187 CrossRef CAS PubMed.
- S. M. Hanash, S. J. Pitteri and V. M. Faca, Nature, 2008, 452, 571–579 CrossRef CAS PubMed.
- K. Blennow, H. Hampel, M. Weiner and H. Zetterberg, Nat. Rev. Neurol., 2010, 6, 131–144 CrossRef CAS PubMed.
- T. A. Addona, S. E. Abbatiello, B. Schilling, S. J. Skates, D. R. Mani, D. M. Bunk, C. H. Spiegelman, L. J. Zimmerman, A.-J. L. Ham, H. Keshishian, S. C. Hall, S. Allen, R. K. Blackman, C. H. Borchers, C. Buck, H. L. Cardasis, M. P. Cusack, N. G. Dodder, B. W. Gibson, J. M. Held, T. Hiltke, A. Jackson, E. B. Johansen, C. R. Kinsinger, J. Li, M. Mesri, T. A. Neubert, R. K. Niles, T. C. Pulsipher, D. Ransohoff, H. Rodriguez, P. A. Rudnick, D. Smith, D. L. Tabb, T. J. Tegeler, A. M. Variyath, L. J. Vega-Montoto, Å. Wahlander, S. Waldemarson, M. Wang, J. R. Whiteaker, L. Zhao, N. L. Anderson, S. J. Fisher, D. C. Liebler, A. G. Paulovich, F. E. Regnier, P. Tempst and S. A. Carr, Nat. Biotechnol., 2009, 27, 633–641 CrossRef CAS PubMed.
- A. Xu, Clin. Chem., 2006, 52, 405–413 CAS.
- O. M. A. El-Agnaf, FASEB J., 2006, 20, 419–425 CrossRef CAS PubMed.
- J. R. Hurst, G. C. Donaldson, W. R. Perera, T. M. A. Wilkinson, J. A. Bilello, G. W. Hagan, R. S. Vessey and J. A. Wedzicha, Am. J. Respir. Crit. Care Med., 2006, 174, 867–874 CrossRef CAS PubMed.
- S. Riethdorf, H. Fritsche, V. Muller, T. Rau, C. Schindlbeck, B. Rack, W. Janni, C. Coith, K. Beck, F. Janicke, S. Jackson, T. Gornet, M. Cristofanilli and K. Pantel, Clin. Cancer Res., 2007, 13, 920–928 CrossRef CAS PubMed.
- E. F. Petricoin, C. Belluco, R. P. Araujo and L. A. Liotta, Nat. Rev. Cancer, 2006, 6, 961–967 CrossRef CAS PubMed.
- P. S. Mitchell, R. K. Parkin, E. M. Kroh, B. R. Fritz, S. K. Wyman, E. L. Pogosova-Agadjanyan, A. Peterson, J. Noteboom, K. C. O'Briant, A. Allen, D. W. Lin, N. Urban, C. W. Drescher, B. S. Knudsen, D. L. Stirewalt, R. Gentleman, R. L. Vessella, P. S. Nelson, D. B. Martin and M. Tewari, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 10513–10518 CrossRef CAS PubMed.
- P. G. Lokhov, M. I. Dashtiev, S. A. Moshkovskii and A. I. Archakov, Metabolomics, 2010, 6, 156–163 CrossRef CAS.
- E. P. Diamandis and D.-E. van der Merwe, Clin. Cancer Res., 2005, 11, 963–965 CAS.
- Y. Qiu, T. H. Patwa, L. Xu, K. Shedden, D. E. Misek, M. Tuck, G. Jin, M. T. Ruffin, D. K. Turgeon, S. Synal, R. Bresalier, N. Marcon, D. E. Brenner and D. M. Lubman, J. Proteome Res., 2008, 7, 1693–1703 CrossRef CAS PubMed.
- R. C. Bollineni, M. Fedorova, M. Blüher and R. Hoffmann, J. Proteome Res., 2014, 13, 5081–5093 CrossRef CAS PubMed.
- G. Engström, L. Stavenow, B. Hedblad, P. Lind, K.-F. Eriksson, L. Janzon and F. Lindgärde, Diabetes, 2003, 52, 442–447 CrossRef.
- G. Engstrom, B. Hedblad, L. Stavenow, P. Lind, L. Janzon and F. Lindgarde, Diabetes, 2003, 52, 2097–2101 CrossRef PubMed.
- G. Engstrom, Circulation, 2002, 105, 2632–2637 CrossRef CAS PubMed.
- G. Engström, B. Hedblad, P. Tydén and F. Lindgärde, Atherosclerosis, 2009, 202, 617–622 CrossRef PubMed.
- P. Lind, B. Hedblad, L. Stavenow, L. Janzon, K. F. Eriksson and F. Lindgärde, Arterioscler., Thromb., Vasc. Biol., 2001, 21, 452–458 CrossRef CAS.
- P. M. Ridker, C. H. Hennekens, J. E. Buring and N. Rifai, N. Engl. J. Med., 2000, 342, 836–843 CrossRef CAS PubMed.
- G. L. Horowitz and B. A. Beckwith, N. Engl. J. Med., 2000, 343, 512–513 CrossRef CAS PubMed.
- A. Festa, R. D'Agostino, R. P. Tracy and S. M. Haffner, Diabetes, 2002, 51, 1131–1137 CrossRef CAS PubMed.
- P. Korantzopoulos, T. Kolettis, K. Siogas and J. Goudevenos, Med. Sci. Monit., 2003, 9, RA225–RA229 CAS.
- J. Danesh, J. G. Wheeler, G. M. Hirschfield, S. Eda, G. Eiriksdottir, A. Rumley, G. D. O. Lowe, M. B. Pepys and V. Gudnason, N. Engl. J. Med., 2004, 350, 1387–1397 CrossRef CAS PubMed.
- G. D. O. Lowe, J. Thromb. Haemostasis, 2005, 3, 1618–1627 CrossRef CAS PubMed.
- http://BiochemDen.com, 2015.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggard, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2050–2055 CrossRef CAS PubMed.
- A. E. Nel, L. Mädler, D. Velegol, T. Xia, E. M. V. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed.
- B. R. Don and G. Kaysen, Semin. Dial., 2004, 17, 432–437 CrossRef PubMed.
- J. Muenchhoff, A. Poljak, A. Thalamuthu, V. B. Gupta, P. Chatterjee, M. Raftery, C. L. Masters, J. C. Morris, R. J. Bateman, A. M. Fagan, R. N. Martins and P. S. Sachdev, Sci. Rep., 2016, 6, 29078 CrossRef CAS PubMed.
- M. Thambisetty, R. Tripaldi, J. Riddoch-Contreras, A. Hye, Y. An, J. Campbell, J. Sojkova, A. Kinsey, S. Lynham, Y. Zhou, L. Ferrucci, D. F. Wong, S. Lovestone and S. M. Resnick, J. Alzheimers Dis., 2010, 22, 1099–1109 CAS.
- C. Song, T. Dinan and B. E. Leonard, J. Affective Disord., 1994, 30, 283–288 CrossRef CAS PubMed.
- A. Hye, S. Lynham, M. Thambisetty, M. Causevic, J. Campbell, H. L. Byers, C. Hooper, F. Rijsdijk, S. J. Tabrizi, S. Banner, C. E. Shaw, C. Foy, M. Poppe, N. Archer, G. Hamilton, J. Powell, R. G. Brown, P. Sham, M. Ward and S. Lovestone, Brain, 2006, 129, 3042–3050 CrossRef CAS PubMed.
- A. ManafiRad, F. Farzadfar, L. Habibi, M. Azhdarzadeh, H. Aghaverdi, K. H. Tehrani, M. Lotfi, P. G. Kehoe, A. Sheidaei, A. Ghasemian, E. R. Darzi, R. Mahmoodi and M. Mahmoudi, J. Alzheimers Dis., 2014, 42, 69–85 CAS.
- A. M. Scott, J. D. Wolchok and L. J. Old, Nat. Rev. Cancer, 2012, 12, 278–287 CrossRef CAS PubMed.
- T. Zheng, N. Pierre-Pierre, X. Yan, Q. Huo, A. J. O. Almodovar, F. Valerio, I. Rivera-Ramirez, E. Griffith, D. D. Decker, S. Chen and N. Zhu, ACS Appl. Mater. Interfaces, 2015, 7, 6819–6827 CAS.
- M. Perfézou, A. Turner and A. Merkoçi, Chem. Soc. Rev., 2012, 41, 2606–2622 RSC.
- Z. Bakhtiary, A. A. Saei, M. J. Hajipour, M. Raoufi, O. Vermesh and M. Mahmoudi, Nanomed. Nanotechnol. Biol. Med., 2016, 12, 287–307 CrossRef CAS PubMed.
- E. Casals, T. Pfaller, A. Duschl, G. J. Oostingh and V. Puntes, ACS Nano, 2010, 4, 3623–3632 CrossRef CAS PubMed.
- J. H. Teichroeb, J. A. Forrest and L. W. Jones, Eur. Phys. J. E, 2008, 26, 411–415 CrossRef CAS PubMed.
- W. Shang, J. H. Nuffer, J. S. Dordick and R. W. Siegel, Nano Lett., 2007, 7, 1991–1995 CrossRef CAS PubMed.
- M. Lundqvist, I. Sethson and B.-H. Jonsson, Langmuir, 2004, 20, 10639–10647 CrossRef CAS PubMed.
- I. Lynch, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2009, 4, 546–547 CrossRef CAS PubMed.
- Z. J. Deng, M. Liang, M. Monteiro, I. Toth and R. F. Minchin, Nat. Nanotechnol., 2011, 6, 39–44 CrossRef CAS PubMed.
- V. K. Lishko, N. P. Podolnikova, V. P. Yakubenko, S. Yakovlev, L. Medved, S. P. Yadav and T. P. Ugarova, J. Biol. Chem., 2004, 279, 44897–44906 CrossRef CAS PubMed.
- R. W. Carrell and B. Gooptu, Curr. Opin. Struct. Biol., 1998, 8, 799–809 CrossRef CAS PubMed.
- K. A. Conway, J. D. Harper and P. T. Lansbury, Biochemistry, 2000, 39, 2552–2563 CrossRef CAS PubMed.
- F. Chiti and C. M. Dobson, Annu. Rev. Biochem., 2006, 75, 333–366 CrossRef CAS PubMed.
- J. W. Kelly, Curr. Opin. Struct. Biol., 1996, 6, 11–17 CrossRef CAS PubMed.
- M. B. Yim, J. H. Kang, H. S. Yim, H. S. Kwak, P. B. Chock and E. R. Stadtman, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 5709–5714 CrossRef CAS.
- M. R. Hayden, S. C. Tyagi, M. M. Kerklo and M. R. Nicolls, J. Pancreas, 2005, 6, 287–302 Search PubMed.
- M. C. Lo Giudice, L. M. Herda, E. Polo and K. A. Dawson, Nat. Commun., 2016, 7, 13475 CrossRef CAS PubMed.
- M. J. Hajipour, J. Raheb, O. Akhavan, S. Arjmand, O. Mashinchian, M. Rahman, M. Abdolahad, V. Serpooshan, S. Laurent and M. Mahmoudi, Nanoscale, 2015, 7, 8978–8994 RSC.
- S. S. An, S.-Y. Wu and J. Hulme, Int. J. Nanomed., 2015, 9 CrossRef PubMed.
- J. H. Shannahan, K. S. Fritz, A. J. Raghavendra, R. Podila, I. Persaud and J. M. Brown, Toxicol. Sci., 2016, 152, 406–416 CrossRef CAS PubMed.
- R. Molinaro, C. Boada, G. M. del Rosal, K. A. Hartman, C. Corbo, E. D. Andrews, N. E. Toledano-Furman, J. P. Cooke and E. Tasciotti, Methodist DeBakey Cardiovasc. J., 2016, 12, 169–174 CrossRef PubMed.
- S. G. Han, B. Newsome and B. Hennig, Toxicology, 2013, 306, 1–8 CrossRef CAS PubMed.
- L. K. Vesterdal, L. Mikkelsen, J. K. Folkmann, M. Sheykhzade, Y. Cao, M. Roursgaard, S. Loft and P. Møller, Toxicol. Lett., 2012, 214, 19–26 CrossRef CAS PubMed.
- C. J. Wingard, D. M. Walters, B. L. Cathey, S. C. Hilderbrand, P. Katwa, S. Lin, P. C. Ke, R. Podila, A. Rao, R. M. Lust and J. M. Brown, Nanotoxicology, 2011, 5, 531–545 CrossRef CAS PubMed.
- A. M. Alkilany, N. N. Mahmoud, F. Hashemi, M. J. Hajipour, F. Farvadi and M. Mahmoudi, Chem. Res. Toxicol., 2016, 29, 943–948 CrossRef CAS PubMed.
- M. Azhdarzadeh, A. A. Saei, S. Sharifi, M. J. Hajipour, A. M. Alkilany, M. Sharifzadeh, F. Ramazani, S. Laurent, A. Mashaghi and M. Mahmoudi, Nanomed., 2015, 10, 2931–2952 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |