Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Micro/nanoelectrochemical probe and chip devices for evaluation of three-dimensional cultured cells
Kosuke
Ino
 *a,
Mustafa
Şen
b,
Hitoshi
Shiku
a and
Tomokazu
Matsue
*c
*a,
Mustafa
Şen
b,
Hitoshi
Shiku
a and
Tomokazu
Matsue
*c
aGraduate School of Engineering, Tohoku University, 6-6-11-406 Aramaki-aza Aoba, Aoba-ku, Sendai 980-8579, Japan. E-mail: kosuke.ino@tohoku.ac.jp
bDepartment of Biomedical Engineering, Izmir Katip Celebi University, 35620 Cigli, Izmir, Turkey
cGraduate School of Environmental Studies, Tohoku University, 6-6-11-604 Aramaki-aza Aoba, Aoba-ku, Sendai 980-8579, Japan. E-mail: matsue@bioinfo.che.tohoku.ac.jp
First published on 26th October 2017
Abstract
Herein, we present an overview of recent research progress in the development of micro/nanoelectrochemical probe and chip devices for the evaluation of three-dimensional (3D) cultured cells. First, we discuss probe devices: a general outline, evaluation of O2 consumption, enzyme-modified electrodes, evaluation of endogenous enzyme activity, and the collection of cell components from cell aggregates are discussed. The next section is focused on integrated chip devices: a general outline, electrode array devices, smart electrode array devices, droplet detection of 3D cultured cells, cell manipulation using dielectrophoresis (DEP), and electrodeposited hydrogels used for fabrication of 3D cultured cells on chip devices are discussed. Finally, we provide a summary and discussion of future directions of research in this field.
1. Introduction
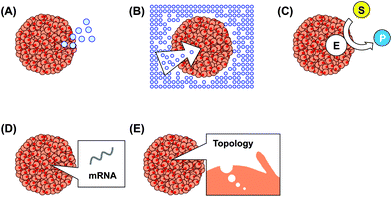
Three-dimensional (3D) cell cultures have received considerable interest in drug discovery, cancer cell biology, stem cell research, and tissue engineering, because unlike 2D cultured cells, 3D cultured cells show in vivo-like cellular responses.1 By culturing cells three-dimensionally, cell–cell interactions, cell–extracellular matrix (ECM) interactions, and cell populations resemble those of in vivo organ tissues. Since 3D cell culture systems provide a simple, fast, and cost-effective cell-based assay tool, such assays are utilized as an alternative to animal testing. For example, tumor spheroids mimicking a native cellular environment are widely used for anti-cancer drug testing.2 In tissue engineering, cells are three-dimensionally cultured to fabricate functional organ tissues. Recently, organ-like structures, termed organoids, have been generated from stem cells using 3D culture systems.3 For these applications, it is necessary to evaluate 3D cultured cells precisely and easily. In addition, single-cell analysis of 3D cultured cells is necessary to investigate the cell population in the culture and the function of individual cells.Several methods have been developed to evaluate cell functions in 3D cultured cells, such as chemical secretion (Fig. 1A), chemical consumption (Fig. 1B), enzyme activity (Fig. 1C), and gene expression (Fig. 1D). In addition, cell density, cell population, morphology, and cell surface topography (Fig. 1E) in 3D cultured cells have been investigated. For the general evaluation of 3D cultured cells, optical methods, such as fluorescence methods, are widely used, because they provide high sensitivity using commercially available reagents. Electrochemical detection has also been proposed as an alternative to fluorescence-based assays, because electrochemical devices show excellent analytical features. For example, the electrochemical method mentioned later is a non-labeling and non-invasive method for the evaluation of respiratory activity in cells. In addition, micro/nanotechnology can provide highly sensitive electrochemical assays and the incorporation of many sensors, among other benefits.
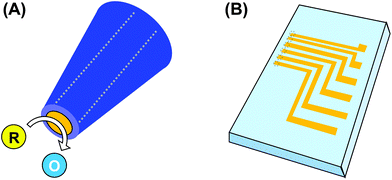
Due to the rapid progress in micro/nanofabrication techniques, micro/nanometer-sized electrodes are easily fabricated. Microelectrodes show several unique features,4 such as high signal-to-noise ratio and electrochemical detection in local areas. Due to these features, micro/nanoelectrochemical devices are widely utilized for evaluation of 3D cultured cells. Electrochemical devices can be roughly categorized into probe and chip devices (Fig. 2). In general, a probe device consists of an electrode covered partially with an insulation layer, and the electrode is exposed at the tip of the device (Fig. 2A). For advanced detection, multi-electrodes and/or multi-capillaries are incorporated into a single probe device. The probe devices are utilized to image local areas in 3D cultured cells and/or collect cell components. Chip devices are fabricated using photolithographic technologies to incorporate electrodes and some microstructures, such as microwells and microfluidics (Fig. 2B). The chip devices are utilized for high-throughput analysis of 3D cultured cells and detection of multiple analytes. In this review, we summarize micro/nanoelectrochemical probe and chip devices used for evaluation of 3D cultured cells.
2. Probe devices
2.1. General outline
Probe devices have been developed for scanning electrochemical microscopy (SECM).5 In SECM, a micro/nanoelectrode is passed over a sample surface to detect local chemical reactions. Although a probe device with a single electrode is generally used for SECM, several types of probe devices have been proposed. For example, multiple electrodes can be incorporated inside or outside a probe device for multi-analyte detection, high-throughput imaging,6 or the combination of working and reference electrodes.7 A glass capillary or dual capillary is used for scanning ion conductance microscopy (SICM).8 When an organic solvent is introduced into a capillary, the probe device is used as a syringe to collect an attoliter-scale volume of solution.9 Double-barreled capillary devices were developed to combine SECM and SICM.10 Field-effect-transistor (FET) sensors with dual carbon nanoelectrodes have been used for ATP detection.11 A flexible probe device has been developed to map target materials on organ tissues.12 Some of these probe devices are utilized for the evaluation of 3D cultured cells.2.2. Evaluation of O2 consumption of 3D cultured cells
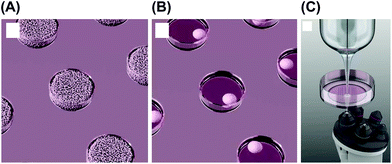
Cell aggregates consume dissolved oxygen, resulting in O2 concentration gradients near the cells. SECM is a powerful tool that can measure O2 concentrations; the probe can detect concentration as a function of position, and the resulting profile allows us to calculate the O2 consumption rate (fmol s−1) of a single cell aggregate.13 In general, a reference electrode should be inserted into a solution in addition to the working electrode (Fig. 3A). However, the process is complicated and unsuitable for measurements of small volumes. To solve these problems, a probe device containing both working and reference electrodes was developed (Fig. 3B).14 To incorporate these electrodes into a single probe device, a Pt pseudo-reference was prepared on the surface of the probe device. The device was successfully applied to the evaluation of O2 consumption of a single embryoid body (EB) from mouse embryonic stem (ES) cells. For simple, array-based detection, cell aggregates were fabricated in a microwell array (Fig. 4).15 In another study, hydrogels containing cells were embedded in silicon microstructures, and a microelectrode was passed above the sample to evaluate cell respiratory activity.16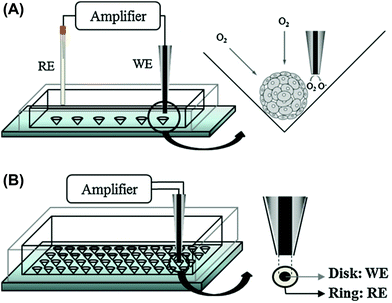 | ||
| Fig. 3 Electrochemical detection of O2 consumption by 3D cultured cells using SECM.14 (A) Conventional SECM system, working (WE) and reference (RE) electrodes are inserted into a solution, and 3D cultured cells are trapped in a microwell. (B) Probe devices containing both working (WE) and reference electrodes (RE) for simplifying the detection procedure. Reproduced with permission from Elsevier ©2012. | ||
 | ||
| Fig. 4 The concept of SECM-based assays for microtissues fabricated in a microwell array.15 (A) Cells are seeded and centrifuged to set them into the microwells. (B) Cell aggregates form after culture. (C) SECM measurements are perfumed for the cell culture arrays. Reproduced from Public Library of Science ©2014. | ||
The effects of glucose-depleted media on O2 consumption of EBs were investigated using SECM.17 According to the results, the differentiated cells showed a stronger adaptation to glucose deprivation than the undifferentiated cells. Furthermore, the relationship between respiratory activity and gene expression in EBs was investigated.18 Many differentiation markers were positively correlated with respiratory activity.
A microwell method19 and the hanging drop culture method20 are widely used to fabricate cell aggregates. The effects of these methods on cell function and density were investigated using SECM. Spheroids of human breast cancer cells (MCF-7) formed in microwells exhibited respiratory activity 2-fold greater than did spheroids produced by the hanging drop method.21
SECM was applied to the evaluation of 3D cultured cells derived from Flk-1(+) and Flk-1(−) cells in EBs comprising mouse ES cells.22 The cell aggregates derived from late stage Flk-1(+) cells showed a relatively small size and low O2 consumption compared to those derived from Flk-1(−) cells.
2.3. Enzyme-modified electrode
Enzyme-modified electrodes have been utilized in selective assays. For example, an electrode modified with lactate oxidase was utilized to monitor the production of lactate by single hepatic spheroids;23 in this report, both the production of lactate and O2 consumption were monitored using an integrated probe device. In another study, glucose oxidase (GOx) was loaded onto a Pt nanoparticle-modified microelectrode to monitor the glucose consumption of pancreatic hamster insulinoma tumor (HIT) spheroids in a self-referencing system.24 The measurements were performed by self-referencing between two different positions to remove large background signals and drift. The probe was used to demonstrate the influence of two different compounds (rotenone and carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP)) on glucose flux around a spheroid.2.4. Evaluation of endogenous enzyme activity of 3D cultured cells
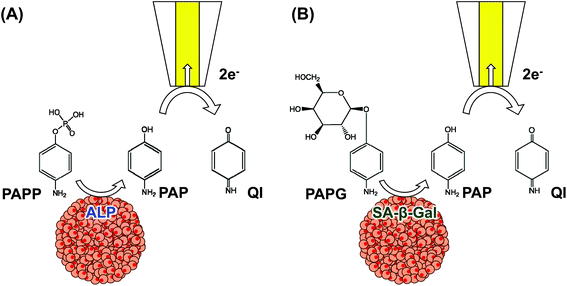
Endogenous ALP is commonly used as an undifferentiated marker of ES cells. SECM was applied as a noninvasive evaluation method to detect ALP activity in EBs (Fig. 5A).14,25,26Additionally, SECM was applied in the non-invasive monitoring of osteogenic differentiation of C2C12 microtissues.27 Differentiation was assessed by measuring the activity of ALP, which is known to be highly expressed during differentiation. SECM was applied to the evaluation of senescence-associated β-galactosidase (SA-β-Gal), which is expressed only in senescent cells28 in MCF-7 spheroids (Fig. 5B).29
Several substrates have been developed for the electrochemical monitoring of endogenous components in cells. For example, caspase-3 activity, an indicator of apoptosis, was detected using enzymatic substrates such as Asp-Glu-Val-Asp-p-nitroaniline DEVD-(pNA)30 and Asp-Glu-Val-Asp-p-methoxyaniline DEVD-(pMA).31pNA and pMA released by the caspase-3 enzyme reaction were electrochemically measured. These substrates will be utilized for the evaluation of 3D cultured cells in the future.
2.5. Collection of cell components from cell aggregates
Double-barreled carbon probes (DBCP) enable the local collection and analysis of cell components (Fig. 6).32,33 Target cells were punctured by an electric pulse between the two electrodes in the DBCP, and the cellular lysates were aspirated thorough the DBCP. The probe device was applied to the evaluation of gene expression in angiogenesis and ES cell-derived cardiomyocytes.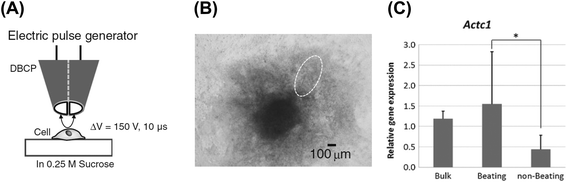 | ||
| Fig. 6 mRNA collection from cells using the DBCP.32 (A) General outline. (B) Phase contrast image of the 3D cultured ES cells differentiated into cardiomyocytes. A dashed line indicates the beating area. (C) Relative gene expression of Actc1 in the cells at the beating area and at the non-beating area. Reproduced with permission from Springer ©2014. | ||
Capillary-type probe devices are widely utilized in SICM detection for imaging at nanometer-scale resolutions. These devices were applied to evaluate an unlabeled secretory protein in living cells34 and nanoscale dynamics of microvilli.35 Although the cell surfaces of 2D cultured cells have not been compared by SICM to those of 3D cultured cells, such comparisons will be performed in the future.
A probe device with two capillaries was used to collect local cell components and image cell surfaces.36 One of the capillaries was filled with an aqueous solution for SICM topographical imaging, and another filled with an organic electrolyte was used as an electrochemical syringe9 to collect cytosol from living cells. The probe device was used to determine positions and collect mRNA. Although the probe device was used only for single-cell analysis in 2D culture, it will be utilized for the evaluation of 3D cultured cells.
A mille-feuille probe allows the lamination of the aqueous and organic phases in a nanopipette under voltage control (Fig. 7).37 The mille-feuille probe was used for continuous collection of different nucleic acid samples and sequential evaluation of gene expression with mRNA barcoding tags. This probe device is useful for multi-gene analysis of 3D cultured cells.
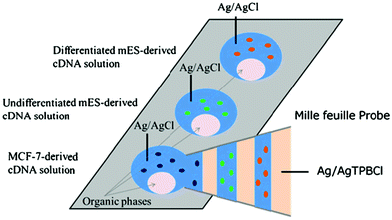 | ||
| Fig. 7 Mille-feuille probe containing the lamination of the aqueous and organic phases under voltage control for simultaneous detection of multiple cDNAs collected with the mille-feuille probe.37 Reproduced with permission from Springer ©2017. | ||
A capillary probe was integrated with an organic electrochemical transistor (OECT) to monitor the integrity of Madin Darby Canine Kidney I (MDCK I) 3D spheroids.38 Briefly, the probe was first used for isolation of relatively small spheroids, and then integrated with an OECT; an Ag/AgCl gate electrode was inserted into the capillary tube. This system allowed recording of electrical resistance through MDCK I spheroids for quality control as well as quantitative toxicity analysis of chemical compounds.
3. Chip devices
3.1. General outline
Chip devices containing multiple electrochemical sensors are useful for high-throughput detection and imaging of 3D cultured cells. In addition, combinations of micro/nanoelectrodes on the devices can provide highly sensitive assays. For example, a comb-type interdigitated microarray (IDA) electrode on a planar device provides a unique system for signal amplification based on redox cycling.39 In redox cycling, two sets of electrodes are located in close proximity to one another. A reacted redox species on the electrodes diffuses and is regenerated on another set of electrodes, so that the electrochemical response can be enhanced. In addition to planar electrodes, nanogap devices with electrode pairs have been proposed for redox cycling-based signal amplification and selective assays.40,41 In addition to electrodes, microwells and microfluidics are incorporated into the devices for trapping cells, manipulating cells, and accumulating target molecules.Several kinds of electrode array devices have been reported for microarray detection,42 and the present review focuses on the chip devices for evaluation of 3D cultured cells.
3.2. Electrode array devices
In addition to SECM probe devices, chip devices have been utilized to evaluate the respiratory activity of single mouse embryos.43–45 In one such device, a single mouse embryo was positioned in a microwell surrounded by electrodes (Fig. 8), and the O2 consumption of the two-cell, morula, and blastocyst stages was successfully measured. In another device, electrodes were arranged 120, 170, 220, 300, 400, and 500 μm from the center of the pit, so that the O2 concentration profile near the cells could be acquired without complicated operations such as scanning with a probe.44 Additionally, cells were 3D cultured directly on flat electrodes for long-term electrochemical monitoring. A screen-printed electrode was incorporated with A549 microtissues in a synthetic hydrogel for a cytotoxicity assay to determine the toxicity of a lung cancer drug.46 Variations in the electrochemical signals acquired by square wave voltammetry in [Fe(CN)6]3− were used in toxicity assessments.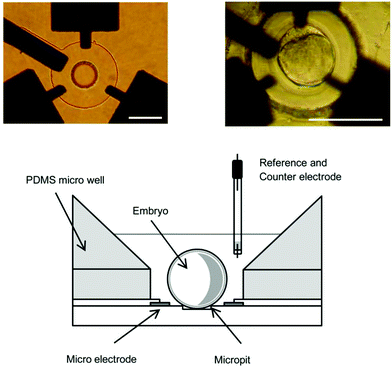 | ||
| Fig. 8 Chip device with three platinum working electrodes, a circular SiO2 insulator layer, and a cylindrical micropit for evaluation of respiratory activity.45 Scale bars: 100 μm. Reproduced with permission from Elsevier ©2011. | ||
To prevent electrode fouling, it is necessary to choose a proper measurement solution. Since serum is adsorbed on an electrode, serum-free medium might be suitable for long-term electrochemical detection. A serum-free solution is commercially available for embryo respiration assay (ERAM-2, Research Institute for the Functional Peptides, Japan). Although the solution contains nutrients such as glucose, there are no chemicals fouling electrodes.
Microfluidics are very useful when different organs are simulated, especially in drug screening. A microfluidic device with an amperometric sensor unit was designed and tested in real-time monitoring of metabolic functions of 3D spheroids, and thus in tracking the dynamics of drug-induced mitochondrial dysfunction.47 The use of an off-chip microfluidic switchboard with pressure-controlled mechanical valves allowed time-resolved sampling and contiguous electrochemical monitoring of glucose uptake and lactate production of multiple HepG2/C3A spheroids over 24 h.
In addition to those using voltammetry, chip devices using electrochemical impedance spectroscopy (EIS) for analysis of 3D microtissues were also reported.48 Because EIS is a non-invasive and label-free method, it has received growing attention for monitoring and evaluating 3D microtissues. In a recent study, multiple electrodes were incorporated into a microfluidic system to construct fully integrated human livers (cancer)- and hearts-on-chips for automated drug screening.49 The surface of the gold microelectrodes were modified with specific antibodies for detection of the liver biomarkers albumin and glutathione S-transferase α (GST-α), as well as the cardiac biomarker creatine kinase MB (CK-MB). The microfluidic system allowed the regeneration of the electrode surface upon saturation with the captured antigens first using H2SO4, and then [Fe(CN)6]3−. The constructed immunobiosensors were successfully used for long-term monitoring of doxorubicin (DOX)- and acetaminophen-induced organ toxicity in a dual-organ platform. In a similar study by the same group, the surface of the gold electrode was functionalized with aptamers specific to CK-MB for monitoring DOX-induced damage to cardiac spheroids in a heart-on-chip platform.50 Successful detection of trace amounts of biomarkers secreted by the spheroids was demonstrated using EIS, and the results were in agreement with the cell viability and beating rate results. A similar sensing platform was also designed for hepatotoxicity assessment and applied to human liver organoids for continuous monitoring of metabolic activity by measuring secreted biomarkers (human albumin and GST-α) for up to 7 days.49 In another study, an interdigitated electrode and microcavity arrays were combined in an impedimetric, real-time measurement system for the comparison of immunotoxin efficacy in 2D and 3D tumor models. The microcavity array comprising 15 cavities with edge lengths of 300–400 μm was successfully used to assess the cytotoxic effects of several immunotoxins on MCF-7 spheroids.51 An optimized design of the microcavity array was later used to construct a 96-well multi-electrode array for impedimetric comparative analysis of the efficacy of several drugs on the same tissue models.52 The multi-electrode array was inserted into a printed circuit board (PCB) comprising 288 electrodes (three electrodes for each well) and 8 counter-electrodes to contact a multiplexer adapter board, to thus individually address each well. The device allowed high-throughput real-time monitoring of the efficacy of DOX and etoposide effects on T98G and SHSY5Y spheroids; the relative therapeutic efficacy was found to be higher for 3D than for 2D tumor models. In addition, a microfluidic device with electrodes embedded into the top and bottom of the microfluidic channel was used for trapping and monitoring MCF-7 microtumor spheroids.53 Spheroids induced changes in the magnitude of the solution resistance consistent with increases in spheroid diameter. This device is suitable for fabrication of multiple channels and electrodes for high-throughput analysis. A similar device with six detection chambers was used by another group for real-time and non-invasive monitoring of human oral cancer cell (OEC-M1) growth on a 3D agarose scaffold.54 The device was then used to monitor the effect of cisplatin, an anti-cancer drug, under continuous perfusion of the medium for 2 days. In addition to drug screening, impedance analysis has also been applied to monitor stem cell differentiation; a significant change in the impedance spectra due to osteogenic differentiation of human mesenchymal stem cells (MSCs) was observed.55 The EIS systems mentioned so far are not suitable for simultaneous monitoring of multiple spheroids over an extended period in an automated fashion. To address the issue, a multiplexed EIS platform was presented in a microfluidic setting.56 This device allowed monitoring the effect of a cancer drug (Fluorouracil) on fifteen HCT116 cancer spheroids simultaneously over four days. Measurements were continuous with a time resolution of 2 min. Due to their low cost and easy fabrication, these devices are very useful for cell analysis. However, it is difficult to incorporate a large number of electrode sensors into a small chip-based device when electrodes are simply arranged. To overcome this difficulty, smart electrode array devices have been proposed.
3.3. Smart electrode array devices
Semiconductor technology has been employed to incorporate multiple electrochemical sensors into a small chip device. For example, we developed a large-scale integration (LSI)-based amperometric sensor for electrochemical imaging, which was designated the Bio-LSI device.57–64 This device was applied to the electrochemical evaluation and real-time monitoring of dopamine release from 3D-cultured PC12 cells62 and ALP activity of EBs.58 As shown in Fig. 9, the chip device allowed us to monitor multiple EBs simultaneously, indicating its utility in high-throughput analysis. In addition, gene silencing by siRNA was monitored to investigate the effects of genes in cell differentiation via the detection of endogenous ALP activity and immunostained sarcomeric α-actinin.59 When the sensor density of the device is low, the distance between sensors and cells affects redox signals. Therefore, the relationship between signals and position should be investigated. A simulation analysis was performed to reveal the positional relationship between the EBs from ES cells and sensor electrodes in order to obtain more precise measurements.61 That study showed that simulation analysis can be applied to the precise electrochemical imaging of 3D cultured cells by normalization of the current signals.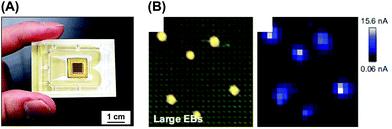 | ||
| Fig. 9 LSI-based electrochemical device. (A) Device image.63 (B) Real-time monitoring of ALP activity of EBs on the device.58 Reproduced with permission from Elsevier ©2013 and 2016. | ||
Consumption or production of compounds by amperometry may affect cell viability. For this reason, potentiometry is superior to amperometry, because redox compounds are not consumed or produced during the detection of open circuit potential. ALP activity of EBs from ES cells was successfully imaged by potentiometry using the LSI device.63
A novel electrochemical approach based the molecular electrochemical switching system was reported as another smart electrode array device.65–71 In this system, two arrays of electrodes (n rows and n columns of electrodes) are orthogonally placed to produce n2 crossing points. By applying proper potential at these electrodes, desired electrochemical reactions can be induced at the target crossing points, such that the crossing points can be used as individual electrochemical sensors. Therefore, n2 electrochemical sensors can be easily incorporated with only 2 n connector pads. This system can be used to measure the ALP activity of ES cells in the evaluation of cell differentiation67 and respiratory activity of MCF-7 spheroids.71
3.4. Droplet detection containing 3D cultured cells
Droplet arrays are useful not only for fabrication of 3D cultured cells, but also for electrochemical detection in compartmented volumes. Several chip devices have been proposed for detection, such as integrated electrode arrays72 and microfluidics.73 Misun et al. reported a droplet array device with enzyme-modified electrodes for body-on-a-chip applications (Fig. 10).73 The microfluidic platform is based on hanging-drop networks for cultivation and analysis of 3D cultured cells. The device monitored both lactate and glucose from single human colon cancer microtissues through amperometry. An EIS readout function was also integrated into the microfluidic hanging-drop platform for microtissue spheroid analysis.74 Briefly, two pairs of microelectrodes were placed directly into the ceiling substrate of each hanging drop to simultaneously control the drop size and the size of spheroids or the beating of cardiac microtissues. The device allowed in situ and continuous monitoring of the growth of cancer spheroids over two days. Compartmented droplets are useful for accumulating target molecules, and evaporation of droplets is utilized for concentrating target molecules. Accumulation of the enzymatic products from EBs of ES cells were monitored during the evaporation process (Fig. 11).66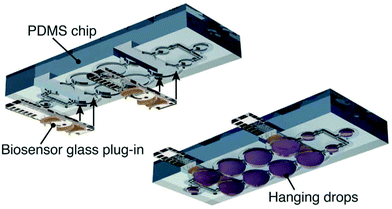 | ||
| Fig. 10 Design of the droplet array microfluidic device with enzyme-modified electrodes.73 Reproduced from Springer Nature ©2016. | ||
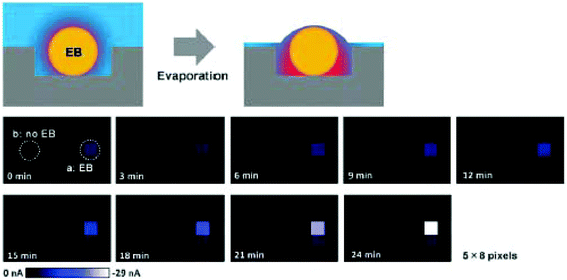 | ||
| Fig. 11 Evaporation-based electrochemical detection of ALP activity in EBs.66 Illustrations of the outline (top row). Time-course analyses of electrochemical images of an EB (middle and bottom row). Redox compounds were accumulated as the incubation time increased. Reproduced from the Royal Society of Chemistry ©2012. | ||
3.5. Cell manipulation using dielectrophoresis (DEP)
DEP was reported by Pohl as the motion of dielectric particles under a non-uniform electric field.75 DEP can be used to manipulate various particles, including uncharged particles. Fig. 12 shows the manipulation of the EBs of ES cells using positive DEP (pDEP).70 As shown in Fig. 12A, an AC electric field was applied between the top and bottom electrodes to guide an EB into the microwell by DEP (Fig. 12B). The excess EBs were flushed away with solution. Since a single EB array was easily fabricated using the DEP technique, this electrochemical system is useful for cell analysis. DEP can also be used to form 3D spheroids.76 Interdigitated, oppositely placed, castellated electrodes were used to pull Jurkat cells into an aggregate via 15 min of pDEP, which allowed the cells to adhere to one another. To demonstrate the reproducibility of the technique, spheroid formation of a different cell line was also shown; in both cases, the cell viability was maintained during aggregation. The technique was later applied to the formation of EBs from ES cells.77 In addition to pDEP, negative DEP (nDEP) was also used for the formation of 3D cell aggregates into arrays.78 In that study, nDEP was employed to gather cells into multiple cell aggregates, which were then encapsulated with a synthetic hydrogel for peeling. Additionally, a microfluidic test system capable of 3D co-culturing with DEP was also reported.79 This microfluidic system featuring cell chambers and interdigitated electrodes was used for the 3D co-culture of hepatocytes and endothelial cells in multiple chambers for liver toxicity assessments.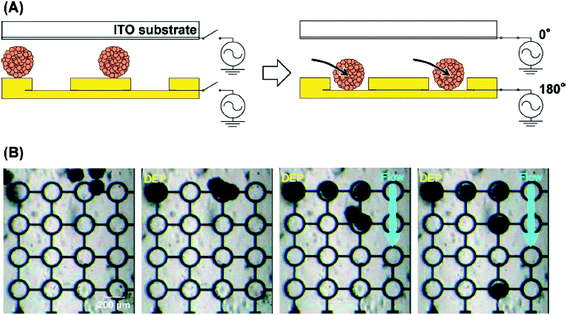 | ||
| Fig. 12 Manipulation of EBs using DEP. (A) Scheme. (B) Optical images of the EBs on the device during DEP and fluidic manipulations.70 Reproduced with permission from the Royal Society of Chemistry ©2015. | ||
3.6. Electrodeposited hydrogels for fabrication of 3D cultured cells on chip devices
An electrochemical device was utilized to fabricate microwell hydrogels, and the microwells were utilized for the 3D culture of cells (Fig. 13).80,81 Acidification-based alginate hydrogel fabrication82 was adapted for the method. Thus, this electrochemical device is useful for preparing 3D cultured cells. In another method using microwells and electrodes, cells were cultured on a self-assembled monolayer (SAM) of alkanethiol.83 The SAM was reductively desorbed from the gold electrodes, allowing 3D cultured cells to be harvested from the microwells.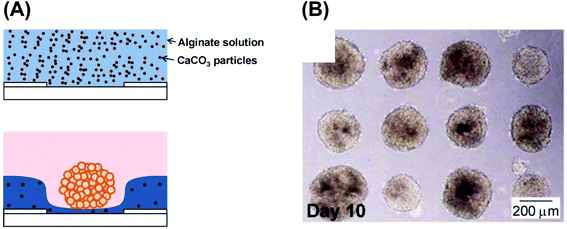 | ||
| Fig. 13 Electrodeposited hydrogel microwells used for 3D cell culturing. (A) Fabrication scheme. (B) HepG2 spheroids in the microwells.81 Reproduced with permission from the Royal Society of Chemistry ©2013. | ||
4. Summary and future directions
In this review, we summarized recent studies on micro/nanoelectrochemical probe and chip devices for evaluation of 3D cultured cells. A summary classifying the target cells and target molecules is shown in Tables 1 and 2. Although several kinds of cells have been evaluated using electrochemical devices as shown in Table 1, it is difficult to discuss which devices/techniques are suitable for each cell type. For high-throughput analysis, electrode array devices and microfluidic devices are suitable. In contrast, probe devices containing a nanoelectrode and a capillary are suitable for imaging of nanometer-sized constructions and collections of cell components, respectively (Table 2). As shown in Table 2, bare electrodes are utilized for evaluation of O2 consumption, endogenous ALP, and dopamine release. In contrast, it is necessary to modify bare electrodes with enzymes for detection of glucose and lactate. Impedimetric analysis is preferred when determining the size of microtissues or constructing immunobiosensors for proteins secreted by microtissues, such as albumin and GST-α. Although it is difficult to compare sensitivity of each technique, chip devices might be suitable for highly sensitive assays, because micro/nanofluidics and micro/nanowells can be easily incorporated into the devices to accumulate and/or amplify redox signals. For example, single molecules were successfully detected using a redox cycling-based nanofluidic device.84,85 As described in another review,42 chip devices will be utilized in organ-on-a-chip systems, which mimic organs in microfluidics.86 Several materials, such as hydrogels and flexible polymers, have been proposed for device fabrication in these applications.| Cell types | Ref. |
|---|---|
| Mouse ES cells | 14, 17, 18, 22, 32, 33, 58, 59, 61, 63, 66–70 and 77 |
| MCF-7 cells | 16, 21, 29, 43, 51, 53 and 71 |
| PC12 cells | 62 |
| Mouse embryos | 44 and 45 |
| Frozen-thawed human embryos | 43 |
| Bro (human melanoma) cells | 2 |
| Pancreatic hamster insulinoma tumor spheroids | 24 |
| C2C12 cells | 27 |
| MDCK I | 38 |
| HepG2/C3A | 47 |
| Rat cardiomyocytes | 2 |
| Human colon carcinoma cell line HCT 116 | 56 and 73 |
| HeLa cells | 15 and 78 |
| Human breast carcinoma cells | 48 |
| HepaRG cells (HPR116) | 23 |
| Human ES cell-derived cardiomyocytes | 50 |
| Jurkat cells | 76 |
| Human myelogenous leukemic cells | 78 |
| Primary human hepatocytes | 49 and 79 |
| Primary human endothelial cells | 79 |
| Human glioblastoma | 52 |
| Human neuroblastoma | 52 |
| Human oral cancer cells | 54 |
| Human lung adenocarcinoma epithelial cells (A549) | 46 |
| Target analytes | Ref. |
|---|---|
| O2 consumption | 13–18, 21–23, 44, 45 and 71 |
| Endogenous ALP | 14, 27, 58, 59, 61, 63 and 66–70 |
| HRP for immunostaining | 59 |
| Glucose | 47 and 73 |
| Lactate | 23, 47 and 73 |
| Electochemical collection and PCR | 32, 33 and 36 |
| Dopamine | 62 |
| SA-β-Gal | 29 |
| Albumin | 49 |
| GST-α | 49 |
| Creatine kinase MB (CK-MB) | 49 and 50 |
| Impedance | 38, 48, 51–56 and 74 |
SICM is being prepared for sales or is commercially available from several companies, e.g. Hokuto Denko (Japan) and HEKA Elektronik (Germany). As described in the section of 2.5, the SICM system was applied to extract cell contents, which was described as an emerging trend for collecting cell components in a perspective.87 Although these methods possess great advantages, further investigations are needed to apply the system for high-aspect-ratio structures. Some of the LSI-based amperometric devices are being prepared for sales, e.g. Bio-LSI (Japan Aviation Electronics Industry, Ltd, Japan) and BioCMOS (BioCMOS Co., Ltd, Japan). For popularizing LSI-based electrochemical devices for bioassays, it is necessary to lower the cost, because these devices are disposable due to electrode fouling during electrochemical detection. Although plasma ashing is effective for cleaning the devices, some modifications are investigated to prevent electrode fouling.
According to a citation analysis, a citation on SICM rapidly increases, and some of the papers focused on bioanalysis, because cell topology can be monitored without cell damage using the technique. There are a few reports focusing on evaluation of 3D cultured cells, but the number of reports will increase. A citation on electrode arrays gradually increases; however, the number of citation to LSI devices for bioanalysis is low. However, the LSI devices will become a trend for cell analysis, because some devices for electrochemical bioanalysis will become commercially available. Notably, integration of microfluidics into such devices is attracting more attention because it facilitates the construction of organ-on-a-chip systems for a variety of applications ranging from toxicity assessment to drug screening.
Although electrochemical methods are useful for evaluation of 3D cultured cells, they must be combined with other methods, such as optical-based image analysis,88,89 to improve their analytical features.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by a Grant-in-Aid for Scientific Research (A) (no. 16H02280), a Grant-in-Aid for Scientific Research (B) (no. 15H035420), and a Grant-in-Aid for Young Scientists (A) (no. 15H05415) from the Japan Society for the Promotion of Science (JSPS). This work was also supported by Special Coordination Funds for Promoting Science and Technology, Creation of Innovation Centers for Advanced Interdisciplinary Research Areas Program from the Japan Science and Technology Agency. This work was also supported by the Asahi Glass Foundation.References
- R. Edmondson, J. J. Broglie, A. F. Adcock and L. Yang, Assay Drug. Dev. Technol., 2014, 12, 207–218 CrossRef CAS PubMed.
- D. Kloss, R. Kurz, H. G. Jahnke, M. Fischer, A. Rothermel, U. Anderegg, J. C. Simon and A. A. Robitzki, Biosens. Bioelectron., 2008, 23, 1473–1480 CrossRef CAS PubMed.
- M. A. Lancaster and J. A. Knoblich, Science, 2014, 345, 1247125 CrossRef PubMed.
- K. Stulik, C. Amatore, K. Holub, V. Marecek and W. Kutner, Pure Appl. Chem., 2000, 72, 1483–1492 CrossRef CAS.
- D. Polcari, P. Dauphin-Ducharme and J. Mauzeroll, Chem. Rev., 2016, 116, 13234–13278 CrossRef CAS PubMed.
- A. Lesch, D. Momotenko, F. Cortes-Salazar, I. Wirth, U. M. Tefashe, F. Meiners, B. Vaske, H. H. Girault and G. Wittstock, J. Electroanal. Chem., 2012, 666, 52–61 CrossRef CAS.
- K. Ino, K. Ono, T. Arai, Y. Takahashi, H. Shiku and T. Matsue, Anal. Chem., 2013, 85, 3832–3835 CrossRef CAS PubMed.
- P. Novak, C. Li, A. I. Shevchuk, R. Stepanyan, M. Caldwell, S. Hughes, T. G. Smart, J. Gorelik, V. P. Ostanin, M. J. Lab, G. W. Moss, G. I. Frolenkov, D. Klenerman and Y. E. Korchev, Nat. Methods, 2009, 6, 279–281 CrossRef CAS PubMed.
- F. O. Laforge, J. Carpino, S. A. Rotenberg and M. V. Mirkin, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 11895–11900 CrossRef CAS PubMed.
- Y. Takahashi, A. I. Shevchuk, P. Novak, Y. Murakami, H. Shiku, Y. E. Korchev and T. Matsue, J. Am. Chem. Soc., 2010, 132, 10118–10126 CrossRef CAS PubMed.
- Y. Zhang, J. Clausmeyer, B. Babakinejad, A. L. Cordoba, T. Ali, A. Shevchuk, Y. Takahashi, P. Novak, C. Edwards, M. Lab, S. Gopal, C. Chiappini, U. Anand, L. Magnani, R. C. Coombes, J. Gorelik, T. Matsue, W. Schuhmann, D. Klenerman, E. V. Sviderskaya and Y. Korchev, ACS Nano, 2016, 10, 3214–3221 CrossRef CAS PubMed.
- T. E. Lin, A. Bondarenko, A. Lesch, H. Pick, F. Cortes-Salazar and H. H. Girault, Angew. Chem., Int. Ed., 2016, 55, 3813–3816 CrossRef CAS PubMed.
- H. Shiku, T. Shiraishi, H. Ohya, T. Matsue, H. Abe, H. Hoshi and M. Kobayashi, Anal. Chem., 2001, 73, 3751–3758 CrossRef CAS PubMed.
- R. Obregon, Y. Horiguchi, T. Arai, S. Abe, Y. Zhou, R. Takahashi, A. Hisada, K. Ino, H. Shiku and T. Matsue, Talanta, 2012, 94, 30–35 CrossRef CAS PubMed.
- A. Sridhar, H. L. de Boer, A. van den Berg and S. Le Gac, PLoS One, 2014, 9, e93618 Search PubMed.
- Y. S. Torisawa, Y. Nashimoto, T. Yasukawa, H. Shiku and T. Matsue, Biotechnol. Bioeng., 2007, 97, 615–621 CrossRef CAS PubMed.
- H. Shiku, N. Aoki, T. Arai, Y. Zhou, K. Y. Inoue, K. Ino and T. Matsue, Electrochemistry, 2016, 84, 302–304 CrossRef CAS.
- H. Shiku, T. Arai, Y. S. Zhou, N. Aoki, T. Nishijo, Y. Horiguchi, K. Ino and T. Matsue, Mol. Biosyst., 2013, 9, 2701–2711 RSC.
- H. C. Moeller, M. K. Mian, S. Shrivastava, B. G. Chung and A. Khademhosseini, Biomaterials, 2008, 29, 752–763 CrossRef CAS PubMed.
- H. Kurosawa, J. Biosci. Bioeng., 2007, 103, 389–398 CrossRef CAS PubMed.
- Y. Zhou, T. Arai, Y. Horiguchi, K. Ino, T. Matsue and H. Shiku, Anal. Biochem., 2013, 439, 187–193 CrossRef CAS PubMed.
- Y. S. Zhou, I. Fujisawa, K. Ino, T. Matsue and H. Shiku, Mol. Biosyst., 2015, 11, 2560–2567 RSC.
- A. Weltin, S. Hammer, F. Noor, Y. Kaminski, J. Kieninger and G. A. Urban, Biosens. Bioelectron., 2017, 87, 941–948 CrossRef CAS PubMed.
- S. K. Jung, J. R. Trimarchi, R. H. Sanger and P. J. Smith, Anal. Chem., 2001, 73, 3759–3767 CrossRef CAS PubMed.
- T. Arai, T. Nishijo, Y. Matsumae, Y. Zhou, K. Ino, H. Shiku and T. Matsue, Anal. Chem., 2013, 85, 9647–9654 CrossRef CAS PubMed.
- M. Sen, Y. Takahashi, Y. Matsumae, Y. Horiguchi, A. Kumatani, K. Ino, H. Shiku and T. Matsue, Anal. Chem., 2015, 87, 3484–3489 CrossRef CAS PubMed.
- A. Sridhar, A. van den Berg and S. Le Gac, Electroanalysis, 2014, 26, 1881–1885 CrossRef CAS.
- M. Storer, A. Mas, A. Robert-Moreno, M. Pecoraro, M. C. Ortells, V. Di Giacomo, R. Yosef, N. Pilpel, V. Krizhanovsky, J. Sharpe and W. M. Keyes, Cell, 2013, 155, 1119–1130 CrossRef CAS PubMed.
- Y. S. Zhou, K. Ino, H. Shiku and T. Matsue, Electrochim. Acta, 2015, 186, 449–454 CrossRef CAS.
- S. Takano, S. Shiomoto, K. Y. Inoue, K. Ino, H. Shiku and T. Matsue, Anal. Chem., 2014, 86, 4723–4728 CrossRef CAS PubMed.
- S. X. Sun, K. Y. Inoue, S. Shiomoto, S. Takano, K. Ino, H. Shiku and T. Matsue, ChemElectroChem, 2017, 4, 941–946 CrossRef.
- Y. Nashimoto, Y. Takahashi, R. Takano, K. Miyashita, S. Yamada, K. Ino, H. Shiku and T. Matsue, Anal. Bioanal. Chem., 2014, 406, 275–282 CrossRef CAS PubMed.
- H. Ito, Y. Nashimoto, Y. S. Zhou, Y. Takahashi, K. Ino, H. Shiku and T. Matsue, Anal. Chem., 2016, 88, 610–613 CrossRef CAS PubMed.
- Y. Nashimoto, Y. Takahashi, H. Ida, Y. Matsumae, K. Ino, H. Shiku and T. Matsue, Anal. Chem., 2015, 87, 2542–2545 CrossRef CAS PubMed.
- H. Ida, Y. Takahashi, A. Kumatani, H. Shiku and T. Matsue, Anal. Chem., 2017, 89, 6015–6020 CrossRef CAS PubMed.
- Y. Nashimoto, Y. Takahashi, Y. S. Zhou, H. Ito, H. Ida, K. Ino, T. Matsue and H. Shiku, ACS Nano, 2016, 10, 6915–6922 CrossRef CAS PubMed.
- H. Ito, M. Tanaka, Y. Zhou, Y. Nashimoto, Y. Takahashi, K. Ino, T. Matsue and H. Shiku, Anal. Bioanal. Chem., 2017, 409, 961–969 CrossRef CAS PubMed.
- M. Huerta, J. Rivnay, M. Ramuz, A. Hama and R. M. Owens, APL Mater., 2015, 3, 030701 CrossRef.
- O. Niwa, M. Morita and H. Tabei, Electroanalysis, 1991, 3, 163–168 CrossRef CAS.
- H. R. Zafarani, K. Mathwig, E. J. R. Sudholter and L. Rassaei, ACS Sens., 2017, 2, 724–728 CrossRef CAS PubMed.
- B. Wolfrum, E. Katelhon, A. Yakushenko, K. J. Krause, N. Adly, M. Huske and P. Rinklin, Acc. Chem. Res., 2016, 49, 2031–2040 CrossRef CAS PubMed.
- K. Ino, H. Shiku and T. Matsue, Curr. Opin. Electrochem. DOI:10.1016/j.coelec.2017.08.004 , in press.
- H. Kurosawa, H. Utsunomiya, N. Shiga, A. Takahashi, M. Ihara, M. Ishibashi, M. Nishimoto, Z. Watanabe, H. Abe, J. Kumagai, Y. Terada, H. Igarashi, T. Takahashi, A. Fukui, R. Suganuma, M. Tachibana and N. Yaegashi, Hum. Reprod., 2016, 31, 2321–2330 CrossRef PubMed.
- K. Hiramoto, M. Yasumi, H. Ushio, A. Shunori, K. Ino, H. Shiku and T. Matsue, Anal. Chem., 2017, 89, 10303–10310 CrossRef CAS PubMed.
- Y. Date, S. Takano, H. Shiku, K. Ino, T. Ito-Sasaki, M. Yokoo, H. Abe and T. Matsue, Biosens. Bioelectron., 2011, 30, 100–106 CrossRef CAS PubMed.
- S. H. Jeong, D. W. Lee, S. Kim, J. Kim and B. Ku, Biosens. Bioelectron., 2012, 35, 128–133 CrossRef CAS PubMed.
- D. Bavli, S. Prill, E. Ezra, G. Levy, M. Cohen, M. Vinken, J. Vanfleteren, M. Jaeger and Y. Nahmias, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E2231–E2240 CrossRef CAS PubMed.
- H. Thielecke, A. Mack and A. Robitzki, Biosens. Bioelectron., 2001, 16, 261–269 CrossRef CAS PubMed.
- S. R. Shin, T. Kilic, Y. S. Zhang, H. Avci, N. Hu, D. Kim, C. Branco, J. Aleman, S. Massa, A. Silvestri, J. Kang, A. Desalvo, M. A. Hussaini, S. K. Chae, A. Polini, N. Bhise, M. A. Hussain, H. Lee, M. R. Dokmeci and A. Khademhosseini, Adv. Sci., 2017, 4, 1600522 CrossRef PubMed.
- S. R. Shin, Y. S. Zhang, D. J. Kim, A. Manbohi, H. Avci, A. Silvestri, J. Aleman, N. Hu, T. Kilic, W. Keung, M. Righi, P. Assawes, H. A. Alhadrami, R. A. Li, M. R. Dokmeci and A. Khademhosseini, Anal. Chem., 2016, 88, 10019–10027 CrossRef CAS PubMed.
- S. Poenick, H. G. Jahnke, M. Eichler, S. Frost, H. Lilie and A. A. Robitzki, Biosens. Bioelectron., 2014, 53, 370–376 CrossRef CAS PubMed.
- M. Eichler, H. G. Jahnke, D. Krinke, A. Muller, S. Schmidt, R. Azendorf and A. A. Robitzki, Biosens. Bioelectron., 2015, 67, 582–589 CrossRef CAS PubMed.
- K. Luongo, A. Holton, A. Kaushik, P. Spence, B. Ng, R. Deschenes, S. Sundaram and S. Bhansali, Biomicrofluidics, 2013, 7, 34108 CrossRef PubMed.
- K. F. Lei, M. H. Wu, C. W. Hsu and Y. D. Chen, Biosens. Bioelectron., 2014, 51, 16–21 CrossRef CAS PubMed.
- C. Hildebrandt, H. Buth, S. Cho, Impidjati and H. Thielecke, J. Biotechnol., 2010, 148, 83–90 CrossRef CAS PubMed.
- S. C. Burgel, L. Diener, O. Frey, J. Y. Kim and A. Hierlemann, Anal. Chem., 2016, 88, 10876–10883 CrossRef CAS PubMed.
- K. Y. Inoue, M. Matsudaira, R. Kubo, M. Nakano, S. Yoshida, S. Matsuzaki, A. Suda, R. Kunikata, T. Kimura, R. Tsurumi, T. Shioya, K. Ino, H. Shiku, S. Satoh, M. Esashi and T. Matsue, Lab Chip, 2012, 12, 3481–3490 RSC.
- M. Sen, K. Ino, K. Y. Inoue, T. Arai, T. Nishijo, A. Suda, R. Kunikata, H. Shiku and T. Matsue, Biosens. Bioelectron., 2013, 48, 12–18 CrossRef CAS PubMed.
- M. Sen, K. Ino, K. Y. Inoue, A. Suda, R. Kunikata, M. Matsudaira, H. Shiku and T. Matsue, Anal. Methods, 2014, 6, 6337–6342 RSC.
- K. Y. Inoue, M. Matsudaira, M. Nakano, K. Ino, C. Sakamoto, Y. Kanno, R. Kubo, R. Kunikata, A. Kira, A. Suda, R. Tsurumi, T. Shioya, S. Yoshida, M. Muroyama, T. Ishikawa, H. Shiku, S. Satoh, M. Esashi and T. Matsue, Lab Chip, 2015, 15, 848–856 RSC.
- Y. Kanno, K. Ino, K. Y. Inoue, A. Suda, R. Kunikata, M. Matsudaira, H. Shiku and T. Matsue, Anal. Sci., 2015, 31, 715–719 CrossRef CAS PubMed.
- H. Abe, K. Ino, C. Z. Li, Y. Kanno, K. Y. Inoue, A. Suda, R. Kunikata, M. Matsudaira, Y. Takahashi, H. Shiku and T. Matsue, Anal. Chem., 2015, 87, 6364–6370 CrossRef CAS PubMed.
- Y. Kanno, K. Ino, C. Sakamoto, K. Y. Inoue, M. Matsudaira, A. Suda, R. Kunikata, T. Ishikawa, H. Abe, H. Shiku and T. Matsue, Biosens. Bioelectron., 2016, 77, 709–714 CrossRef CAS PubMed.
- K. Ino, Y. Kanno, K. Y. Inoue, A. Suda, R. Kunikata, M. Matsudaira, H. Shiku and T. Matsue, Angew. Chem., Int. Ed., 2017, 56, 6818–6822 CrossRef CAS PubMed.
- K. Ino, W. Saito, M. Koide, T. Umemura, H. Shiku and T. Matsue, Lab Chip, 2011, 11, 385–388 RSC.
- K. Ino, Y. Kanno, T. Nishijo, T. Goto, T. Arai, Y. Takahashi, H. Shiku and T. Matsue, Chem. Commun., 2012, 48, 8505–8507 RSC.
- K. Ino, T. Nishijo, T. Arai, Y. Kanno, Y. Takahashi, H. Shiku and T. Matsue, Angew. Chem., Int. Ed., 2012, 51, 6648–6652 CrossRef CAS PubMed.
- K. Ino, T. Nishijo, Y. Kanno, F. Ozawa, T. Arai, Y. Takahashi, H. Shiku and T. Matsue, Electrochemistry, 2013, 81, 682–687 CrossRef CAS.
- K. Ino, Y. Kanno, T. Nishijo, H. Komaki, Y. Yamada, S. Yoshida, Y. Takahashi, H. Shiku and T. Matsue, Anal. Chem., 2014, 86, 4016–4023 CrossRef CAS PubMed.
- Y. Kanno, K. Ino, H. Shiku and T. Matsue, Lab Chip, 2015, 15, 4404–4414 RSC.
- K. Ino, Y. Yamada, Y. Kanno, S. Imai, H. Shiku and T. Matsue, Sens. Actuators, B, 2016, 234, 201–208 CrossRef CAS.
- H. Zhang, T. Oellers, W. Feng, T. Abdulazim, E. N. Saw, A. Ludwig, P. A. Levkin and N. Plumere, Anal. Chem., 2017, 89, 5832–5839 CrossRef CAS PubMed.
- P. M. Misun, J. Rothe, Y. R. F. Schmid, A. Hierlemann and O. Frey, Microsyst. Nanoeng., 2016, 2, 16022 CrossRef CAS.
- Y. R. F. Schmid, S. C. Burgel, P. M. Misun, A. Hierlemann and O. Frey, ACS Sens., 2016, 1, 1028–1035 CrossRef CAS.
- H. A. Pohl, J. Appl. Phys., 1951, 22, 869–871 CrossRef CAS.
- A. Sebastian, A. M. Buckle and G. H. Markx, Biotechnol. Bioeng., 2007, 98, 694–700 CrossRef CAS PubMed.
- S. Agarwal, A. Sebastian, L. M. Forrester and G. H. Markx, Biomicrofluidics, 2012, 6, 24101–2410111 CrossRef PubMed.
- R. G. Abdallat, A. S. Ahmad Tajuddin, D. H. Gould, M. P. Hughes, H. O. Fatoyinbo and F. H. Labeed, Electrophoresis, 2013, 34, 1059–1067 CrossRef CAS PubMed.
- J. Schutte, B. Hagmeyer, F. Holzner, M. Kubon, S. Werner, C. Freudigmann, K. Benz, J. Bottger, R. Gebhardt, H. Becker and M. Stelzle, Biomed. Microdevices, 2011, 13, 493–501 CrossRef PubMed.
- F. Ozawa, K. Ino, H. Shiku and T. Matsue, Materials, 2016, 9, 744 CrossRef PubMed.
- F. Ozawa, K. Ino, T. Arai, J. Ramon-Azcon, Y. Takahashi, H. Shiku and T. Matsue, Lab Chip, 2013, 13, 3128–3135 RSC.
- Y. Cheng, X. L. Luo, J. Betz, G. F. Payne, W. E. Bentley and G. W. Rubloff, Soft Matter, 2011, 7, 5677–5684 RSC.
- R. Inaba, A. Khademhosseini, H. Suzuki and J. Fukuda, Biomaterials, 2009, 30, 3573–3579 CrossRef CAS PubMed.
- S. G. Lemay, S. Kang, K. Mathwig and P. S. Singh, Acc. Chem. Res., 2013, 46, 369–377 CrossRef CAS PubMed.
- P. S. Singh, E. Katelhon, K. Mathwig, B. Wolfrum and S. G. Lemay, ACS Nano, 2012, 6, 9662–9671 CrossRef CAS PubMed.
- S. N. Bhatia and D. E. Ingber, Nat. Biotechnol., 2014, 32, 760–772 CrossRef CAS PubMed.
- S. G. Higgins and M. M. Stevens, Science, 2017, 356, 379–380 CrossRef CAS PubMed.
- R. Kato, M. Matsumoto, H. Sasaki, R. Joto, M. Okada, Y. Ikeda, K. Kanie, M. Suga, M. Kinehara, K. Yanagihara, Y. Liu, K. Uchio-Yamada, T. Fukuda, H. Kii, T. Uozumi, H. Honda, Y. Kiyota and M. K. Furue, Sci. Rep., 2016, 6, 34009 CrossRef CAS PubMed.
- R. Nagasaka, Y. Gotou, K. Yoshida, K. Kanie, K. Shimizu, H. Honda and R. Kato, J. Biosci. Bioeng., 2017, 123, 642–650 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |