Open Access Article
Open Access ArticleSpecificity of SNP detection with molecular beacons is improved by stem and loop separation with spacers†
Valentina M.
Farzan
ab,
Mikhail L.
Markelov
c,
Alexander Yu.
Skoblov
d,
German A.
Shipulin
b and
Timofei S.
Zatsepin
 *abe
*abe
aSkolkovo Institute of Science and Technology, 3 Nobel Street, Innovation Center “Skolkovo”, 143026 Skolkovo, Russia. E-mail: t.zatsepin@skoltech.ru; Fax: +(7) 495-939-5411; Tel: +(7) 495-939-5411
bCentral Research Institute of Epidemiology, Novogireevskaya 3a, 111123 Moscow, Russia
cResearch Institute of Occupational Health, Prospekt Budenogo 31, 105275 Moscow, Russia
dMetkinen Chemistry Oy., Microkatu 1, MicroTower Part-M, FI-70210 Kuopio, Finland
eDepartment of Chemistry, Lomonosov Moscow State University, Leninskie Gory 1-3, 119992 Moscow, Russia
First published on 13th February 2017
Abstract
Molecular beacons (MBs) are valuable tools in molecular biology, clinical diagnostics and analytical chemistry. Here we describe a novel approach for the design of MBs with nucleotide or non-nucleotide linkers between the stem and loop regions. Such modified MBs have significantly improved specificity and performance for single nucleotide polymorphism (SNP) detection. These advantages are especially distinct, when compared to the classic MBs, in the case of possible interactions between the stem and loop regions. We demonstrated the applicability of such modified MBs for the discrimination of common Factor V, NOS3 and ADRB2 SNPs in model plasmids and in clinical samples. The developed approach could be applicable not only to fluorescently labeled MBs, but also to other biosensors based on nucleic acids with stem–loop structures.
Introduction
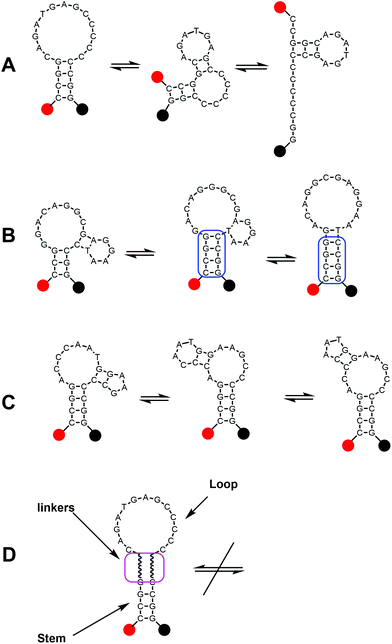
Molecular beacons (MBs) are valuable tools in molecular biology, clinical diagnostics and analytical chemistry.1 They are widely used as reporters in various PCR techniques and for mRNA or microRNA detection in vitro.2 The first MBs were developed for targeting complementary nucleic acids,3 but a number of methods for detection of the activity of DNA-interacting enzymes,4 metal ions,5 toxins and other analytes6 based on MBs were developed later. In the beginning, MBs were designed in a simple stem–loop pattern; but soon evolved into more complicated structures, including di-, tri-, tetra- and even penta-component variants.7 In addition, methods of enzyme-assisted signal amplification exist for MB based detection and these methods significantly improve sensitivity of the assay without affecting specificity.8 Until now, the most common targets for MBs are single nucleotide polymorphisms (SNP), that are the most frequent mutations in the genome and some of them are associated with genetic disorders.9The extraordinary selectivity of MBs for SNPs in complementary nucleic acids is based on the equilibrium between closed and opened forms.10 This equilibrium can be tuned by sequence changes in the stem and loop structures, improving either sensitivity or specificity of SNP detection. However, this approach is not universal and in some cases worsens SNP discrimination by MBs due to nonspecific target recognition. Another common problem in MB design is the formation of multiple secondary structures that compete with each other, including disruption of the stem–loop structure (Fig. 1). In some cases, this leads to the increase of background fluorescence or to poor performance of the probes due to the participation of neighboring nucleotides from the loop in the stem formation (Fig. 1A and B). Another related problem is that common G/A or C/T substitutions are tricky to differentiate by MBs. Indeed, guanine can form a pair with both complementary cytosine and thymine in a DNA duplex with a minimal difference in stability. These interactions can lead to improper probe hybridization. As a result, one can observe an increase of fluorescence for both probes in the assay11 that can give discordant results. In these cases short MB probes conjugated with minor groove binders (MGB) are used to improve specificity for A/G discrimination.12 MGB residues significantly improve duplex stability without affecting specificity. However MGB residues also increase the cost of the probe and of the whole assay. Several studies on MB improvement were directed to modifying the stem regions in MBs to exclude stem participation in hybridization with DNA targets (stem invasion) as this effect results in high background signals and poor analytical sensitivity.13 The introduction of inverted 3′–5′ sequences,13aL-nucleotides13b or modified nucleotides,13c–e to the stem region lead to a significant improvement of MB selectivity. However these variants are not widely used due to high costs or complicated synthesis of modified residues.
In this study we demonstrate that a simple separation of stem and loop parts in MBs by commercially available spacers (hexaethylene glycol or three 2′-deoxyinosines) significantly improves the specificity of SNP detection. Oligo ethylene glycol spacers are widely used in the design of modified DNA.14 Spacers in synthetic oligonucleotides are used to improve the sensitivity of solid phase assays based on immobilized oligonucleotides,14a to simplify the assembly of complex DNA nanostructures,14b,c to block polymerase bypassing of scorpion probes in qPCR14d and to modulate DNA binding properties.14e It was shown previously that the introduction of oligo ethylene glycol linkers into oligonucleotides results in an independent thermodynamic behavior of connected parts.15 We conclude that separation of the stem and loop in molecular beacons should result in better performance of the probes and simplify MB design due to stem fixation (Fig. 1D). As a result of incorporation of spacers into MBs the stem part is unable to participate in the formation of joint structures with the loop part like the one presented in Fig. 1B. Prevention of such undesired interactions significantly improves MB applicability and simplifies MB design. We also propose the use of the universal CCGG stem sequence for MBs as the sole G/C content provides high thermal stability while two adjacent guanine residues opposite to the fluorescent dye provide additional quenching with a minimal influence on the final fluorescence of MBs. Thus this stem sequence could be used as a universal one for all probes in the assay. Also such uniformity can simplify the design of MBs for common SNP targets.
Materials and methods
Oligonucleotide synthesis, purification and characterization
Oligonucleotides were synthesized on the ABI 3400 synthesizer by the phosphoramidite method, according to the manufacturer's recommendations. Protected 2′-deoxyribonucleoside 3′-phosphoramidites, Unylinker-CPG (500 Å) and S-ethylthio-1H-tetrazole were purchased from ChemGenes; 5′-FAM phosphoramidite from Primetech LLC; 5′-SIMA phosphoramidite, BHQ1 CPG, 2′-deoxyinosine and hexaethylene glycol phosphoramidites from Glen Research. All oligonucleotides were cleaved from the support and deprotected using concentrated aqueous ammonia for 8 h at 55 °C. Primers were precipitated with 4% lithium perchlorate in acetone and used without further purification after confirmation of the molecular mass by LC-MS. Oligonucleotide probes were purified by denaturing PAGE followed by RP-HPLC. The conditions of PAGE, HPLC purifications and analysis were the same as previously published.16 ESI-MS spectra for oligonucleotides were recorded using a Bruker Maxis Impact q-TOF system as described earlier (Table S2†).qPCR
Quantitative PCR was performed using primer pairs (Table S1†) and the template plasmid pGem®-t (Promega) containing cloned fragments of the genes (primer positions are underlined; the probe sequence is highlighted in bold): ADRB2 (166 bp)![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif) gactgcgcgccatggggcaac ccgggaacggcagcgccttcttgctggcacccaat
gactgcgcgccatggggcaac ccgggaacggcagcgccttcttgctggcacccaat gaagccatgcgccggaccacgacgtc acgcaggaaagggacgaggtgtgggtggtgggcatgggcatcg
gaagccatgcgccggaccacgacgtc acgcaggaaagggacgaggtgtgggtggtgggcatgggcatcg![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) ; NOS3 (140 bp)
; NOS3 (140 bp) ![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) ggctggaccccaggaaa cggtcgcttcgacgtgctgcccctgctgctgcaggccccagatga
ggctggaccccaggaaa cggtcgcttcgacgtgctgcccctgctgctgcaggccccagatga cccccagaac tcttccttctgccccccgagctggtc
cccccagaac tcttccttctgccccccgagctggtc![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif) ; FV (127 bp)
; FV (127 bp) ![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif) tgtaagagcagatccctg gacaggc
tgtaagagcagatccctg gacaggc aggaatacaggtattttgtccttgaagtaacctttcagaaattctgagaattt
aggaatacaggtattttgtccttgaagtaacctttcagaaattctgagaattt![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[c with combining low line]](https://www.rsc.org/images/entities/char_0063_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[g with combining low line]](https://www.rsc.org/images/entities/char_0067_0332.gif) . Plasmid concentration was estimated by limiting dilution analysis using qPCR with the pUC/M13 Reverse and pUC/M13 Forward Sequencing Primer® (Promega) in the presence of 1× Evagreen® dye (Biotium). The reaction mixtures contained 5 μL PCR-mix-2-FRT (AmpliSens, Russia), 2.5 U of TaqF polymerase (AmpliSens, Russia), 176 μM dNTPs (Biosan, Russia), 1× Evagreen® dye (Biotium) and 0.36 μM of each primer; total reaction volume 25 μL.
. Plasmid concentration was estimated by limiting dilution analysis using qPCR with the pUC/M13 Reverse and pUC/M13 Forward Sequencing Primer® (Promega) in the presence of 1× Evagreen® dye (Biotium). The reaction mixtures contained 5 μL PCR-mix-2-FRT (AmpliSens, Russia), 2.5 U of TaqF polymerase (AmpliSens, Russia), 176 μM dNTPs (Biosan, Russia), 1× Evagreen® dye (Biotium) and 0.36 μM of each primer; total reaction volume 25 μL.
The samples of the pGem®-t plasmid with the NOS3 fragment or the ADRB2 fragment or the Factor V fragment were mixed with 5 μL PCR-mix-2-FRT (AmpliSens, Russia), 2.5 U of TaqF polymerase (AmpliSens, Russia), 176 μM dNTPs (Biosan, Russia); the total reaction volume was 25 μL. The final concentrations of the forward and reverse primers and fluorescently labeled probes were 0.04, 0.36 and 0.12 μM, respectively. The amplification reactions were performed in the RotorGene 6000 (Corbett Research) under the following cycling conditions: 0th cycle at 95 °C/15 min; then 50 cycles at 95 °C/10 s, 47 °C/20 s and 72 °C/20 s. The fluorescence measurements were recorded at the end of the detection step (47 °C) during each of the 50 cycles. The experiments were repeated twice and data were calculated using the Corbett Research software.
We used DNA samples from patients that were already analyzed for SNPs during routine studies in CRIE by pyrosequencing assay.17 Our studies were approved by the CRIE ethics committee as the samples were anonymized and no data from patients were used. DNA for all experiments was extracted from blood with the Ribo-prep kit (Ecoli s.r.o., Slovakia) according to the manufacturer's recommendations. The DNA was dissolved in MilliQ water (50 μL) and aliquots were stored at −70 °C. After confirmation of the genotype, 50–100 ng DNA from the sample were analyzed as mentioned above using LATE-qPCR.22
Results and discussion
In this study we used three common SNPs: NOS3 (G894T, rs1799983), associated with the risk of cardiovascular diseases18 and hypertension; Factor V also known as the Leiden mutation (A1601G, rs6025), which increases the risk of venous thromboembolism19 and ADRB2 (A285G, rs1042713) has an influence on blood pressure and the risk of asthma.20 We have chosen two A/G and one G/T SNP to study if our approach can be universal for various SNP types. Another reason for this choice was the relatively high occurrence of homozygosis for both NOS3 and ADRB2 in the human population, which simplifies the collection of clinical samples.First, we developed the classic MBs that were able to discriminate NOS3, ADRB2 and FV SNP during linear amplification (Table 1). We used a novel SIMA dye for the HEX channel as it proved to be more robust during oligonucleotide synthesis, deprotection and purification in comparison to the common HEX dye.21 The R6G dye,16 another alternative to HEX, increased the melting temperature of MB and MB duplexes with the target, thus complicating the design of MB pairs (data not shown). Therefore SIMA was found to be the optimal choice for MB synthesis and application for SNP detection.
| Name | Sequence (5′3′) |
|---|---|
a Structures of linkers: 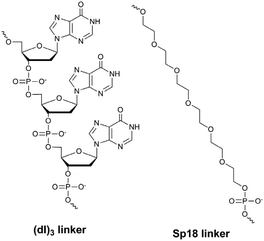
|
|
| b0-adrb2-Fam | Fam-ccggaccaatggaagcccccgg-BHQ1 |
| b0-adrb2-Hex | SIMA-ccggacccaatagaagccaccgg-BHQ1 |
| b1-adrb2-Fam |
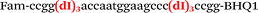
|
| b1-adrb2-Hex |
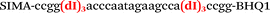
|
| b2-adrb2-Fam |
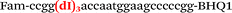
|
| b2-adrb2-Hex |
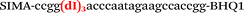
|
| b3-adrb2-Fam |
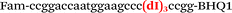
|
| b3-adrb2-Hex |
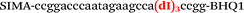
|
| b4-adrb2-Fam |
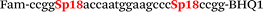
|
| b4-adrb2-Hex |
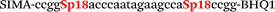
|
| b5-adrb2-Fam |
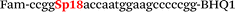
|
| b5-adrb2-Hex |
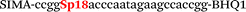
|
| b6-adrb2-Fam |
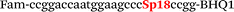
|
| b6-adrb2-Hex |
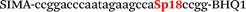
|
| b0-nos3-Fam | Fam-ccggcagatgagcccccccgg-BHQ1 |
| b0-nos3-Hex | SIMA-ccggcagatgatccccaccgg-BHQ1 |
| b1-nos3-Fam |
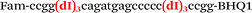
|
| b1-nos3-Hex |
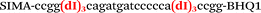
|
| b2-nos3-Fam |
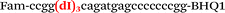
|
| b2-nos3-Hex |
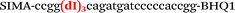
|
| b3-nos3-Fam |
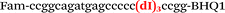
|
| b3-nos3-Hex |
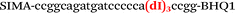
|
| b4-nos3-Fam |
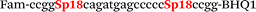
|
| b4-nos3-Hex |
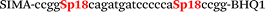
|
| b5-nos3-Fam |
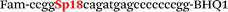
|
| b5-nos3-Hex |
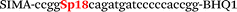
|
| b6-nos3-Fam |
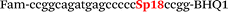
|
| b6-nos3-Hex |
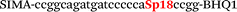
|
| b0-FV-Fam | Fam-ccgggacaggcgaggaatccgg-BHQ1 |
| b0-FV-Hex | SIMA-ccgggacaggcaaggaataccgg-BHQ1 |
| b1-FV-Fam |
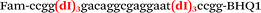
|
| b1-FV-Hex |
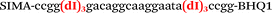
|
| b2-FV-Fam |
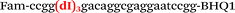
|
| b2-FV-Hex |
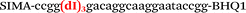
|
| b3-FV-Fam |
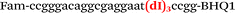
|
| b3-FV-Hex |
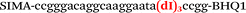
|
| b4-FV-Fam |
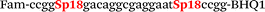
|
| b4-FV-Hex |
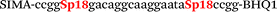
|
| b5-FV-Fam |
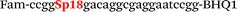
|
| b5-FV-Hex |
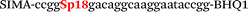
|
| b6-FV-Fam |
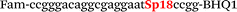
|
| b6-FV-Hex |
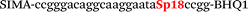
|
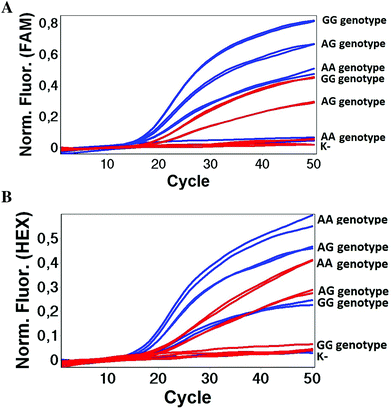
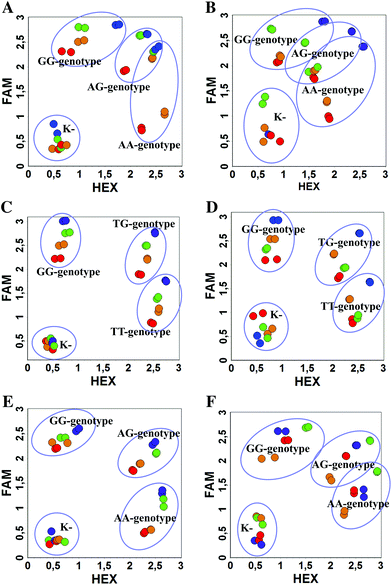
We used the LATE PCR technique,22 based on regular PCR, followed by linear amplification of ssDNA. This approach increases the final fluorescence of MBs, due to the absence of competition during hybridization with the complementary strand that takes place in the case of dsDNA after regular PCR amplification. LATE PCR is more robust and universal than conventional asymmetric PCR. In all cases, we observed a significant nonspecific increase of fluorescent signals for the classic MBs during SNP discrimination in the case of non-complementary base pairs (for Factor V – Fig. 2, for NOS3 and ADRB2 – Fig. S1 and S2†). The same problem was found for all SNPs tested in this study. To prove the usefulness of stem–loop separation, we synthesized a number of modified MBs. We introduced nucleotide (three 2′-deoxyinosines) and non-nucleotide (hexaethylene glycol or Sp18) linkers between the stem and loop parts of MBs (Table 1). These linkers were inserted either only into 5′- or into 3′-stem–loop transitions in MBs or into both 5′- and 3′-transitions (Table 1). Such modifications notably improved the selectivity of MBs (Fig. 2, red series; Fig. 3). Only a minimal increase of fluorescence was observed for mismatched G–T and A–C pairs with modified MBs in comparison to the classic MBs (Fig. 2, red series). However, the introduction of linkers leads to some increase of background fluorescence, probably due to the distortion of the MB structure, which resulted in a decreased final signal (Fig. 2). However genotyping is commonly carried out by the analysis of a relative increase of fluorescence across the FAM and HEX channels during DNA amplification, and represented as a scatter plot (Fig. 3). Unlike allelic discrimination, the determination of the genotype is made on the basis of regions defined on the scatter plot rather than a single threshold or final fluorescence. Therefore a decreased fluorescent output does not influence the robustness of the assay.
As can be seen in Fig. 3, there is no doubt that the probability of heterozygote discrimination is significantly increased for modified MBs in comparison to the classic ones (Fig. 3). In general, MBs simultaneously modified at the 5′- and 3′-ends or only at the 5′ end gave better results, while 3′-end modifications only did not improve the SNP discrimination properties of the probes. The effect was more pronounced for Factor V and NOS3 SNPs while all modified variants of the ADRB2 SNP were more comparable with the classic MBs. We analyzed possible structures for the MBs predicted by using the Mfold software23 (Fig. 1). In the case of Factor V and NOS3 MBs, the 5′-end of the stem is involved in the formation of all alternative structures. Separation of the stem and loop regions by the linker fixes the optimal structure. For ADRB2 MBs, alternative structures are formed in the loop region internally and therefore, the introduced modifications do not significantly improve the probe.
By comparing the results for hexaethylene glycol (Sp18) (Fig. 3A, C, and E) and triple dI (Fig. 3B, D, and F) linkers, we can conclude that a Sp18 linker provides a more pronounced effect on MB applicability to SNP detection. Hexaethylene glycol is rather flexible, while 2′-deoxyinosines form a more rigid linkage in the MBs due to stacking interactions between heterocycles. Also, the hypoxanthine moiety is a universal base, as it can form Watson–Crick pairs with all four natural bases.24 However, the stability of such pairs is lower than that of native base pairs, especially with guanine, and is even lower for self-pairing. Therefore even if 2′-deoxyinosines would be able to interact with the loop region, such structures will be rather unstable and should not influence the functioning of probes. Thus, we have chosen a Sp18 linker for further applications.
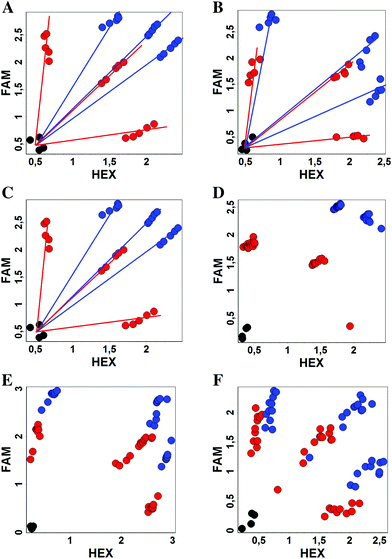
We studied the dependence of the signal distribution in scatterplot analysis on the plasmid target concentration as the amount of DNA in clinical samples can vary significantly. We varied the concentration of all plasmid DNA targets in wide intervals – 1 × 105–1 × 108 copies per mL. In all cases, the signal intensity decreased for modified MBs but the resolution of the SNP by scatterplot analysis was comparable (Fig. 4A–C). For all modified MBs, SNP selectivity was better than the classic MBs. With the classic MBs, cross-signaling was increased in the samples with the highest concentration of DNA, while it remained minimal for modified MBs and minor concentration dependence was observed.
To demonstrate the practical utility of modified MBs, we studied DNA samples from patients that were already analyzed for SNPs during routine studies in CRIE. Based on these data we examined previously analyzed clinical samples for Factor V, NOS3, and ADRB2 SNPs (Fig. 4D–F). In all cases, the genotype was correctly confirmed both by the classic and modified MBs. However, in the case of FV and ADRB2, SNP hetero- and homozygotes are discriminated less obviously by the classic MBs while the modified MBs give more significant differences. Also one sample with the ADRB2 heterozygote contained a rather low amount of DNA but was nevertheless predicted correctly by the analysis.
Conclusions
We have demonstrated that the incorporation of inexpensive nucleotide or non-nucleotide linkers between the stem and loop regions in molecular beacons during automated oligonucleotide synthesis significantly improves their specificity and performance in SNP discrimination. Incorporation of linkers dissects the stem and loop regions in MBs, eliminates some non-specific internal interactions and improves the quality of the analysis. We demonstrated that such modified molecular beacons outperform the classic MBs for ADRB2, NOS3 and Factor V SNPs. Analysis by scatterplot of clinical samples demonstrated that these MBs are more robust for the determination of A/G substitutions than the classic MBs. Previously developed approaches13 that restrict undesired stem interactions were focused mainly on the decrease of stem invasion or interactions of the stems with DNA targets. Here, we develop an approach that not only solves the problem of stem invasion due to the independent thermodynamic behavior of hexaethylene glycol separated parts, but also excludes stem–loop interactions, which increase the background and decrease efficiency of the assay. Excellent SNP selectivity, simple synthesis and moderate cost of the MBs developed in this study exceed the previously developed MBs. There is no doubt that our approach is applicable not only to fluorescently labeled MBs, but also to other biosensors based on the use of nucleic acids with stem–loop structures.Acknowledgements
This work has been supported by the Russian Science Foundation (project no. 14-24-00061 – oligonucleotide synthesis). We are grateful to Ms E. Dunaeva (CRIE) for providing samples of human DNA with validated SNPs and Ms D. Leboeuf (Skoltech) for critical reading of the manuscript.Notes and references
- (a) K. Wang, Z. Tang, C. J. Yang, Y. Kim, X. Fang, W. Li, Y. Wu, C. D. Medley, Z. Cao, J. Li, P. Colon, H. Lin and W. Tan, Angew. Chem., Int. Ed., 2009, 48, 856 CrossRef CAS PubMed; (b) Y. Li, X. Zhou and D. Ye, Biochem. Biophys. Res. Commun., 2008, 373, 457 CrossRef CAS PubMed; (c) K. Huang and A. A. Marti, Anal. Bioanal. Chem., 2012, 402, 3091 CrossRef CAS PubMed.
- (a) A. Giannetti, S. Tombelli and F. Baldini, Anal. Bioanal. Chem., 2013, 405, 6181 CrossRef CAS PubMed; (b) J. H. Lee, J. A. Kim, M. H. Kwon, J. Y. Kang and W. J. Rhee, Biomaterials, 2015, 54, 116 CrossRef CAS PubMed.
- (a) S. Tyagi and F. R. Kramer, Nat. Biotechnol., 1996, 14, 303 CrossRef CAS PubMed; (b) S. Tyagi, D. P. Bratu and F. R. Kramer, Nat. Biotechnol., 1998, 16, 49 CrossRef CAS PubMed.
- (a) L. Zhang, T. Hou, H. Li and F. Li, Analyst, 2015, 140, 4030 RSC; (b) C. A. Belon and D. N. Frick, BioTechniques, 2008, 45, 433 CrossRef CAS PubMed.
- (a) H. Xu, X. Zhu, H. Ye, L. Yu, X. Liu and G. Chen, Chem. Commun., 2011, 47, 12158 RSC; (b) L. Zhang, J. X. Wong, X. Li, Y. Li and H. Z. Yu, Anal. Chem., 2015, 87, 5062 CrossRef CAS PubMed; (c) S. Zhan, Y. Wu, L. Wang, X. Zhan and P. Zhou, Biosens. Bioelectron., 2016, 86, 353 CrossRef CAS PubMed.
- (a) C. Ma, Z. Tang, K. Wang, X. Yang and W. Tan, Analyst, 2013, 138, 3013 RSC; (b) Y. Wang, Q. Sun, L. Zhu, J. Zhang, F. Wang, L. Lu, H. Yu, Z. Xu and W. Zhang, Chem. Commun., 2015, 51, 7958 RSC; (c) S. M. Sanzani, M. Reverberi, C. Fanelli and A. Ippolito, Toxins, 2015, 7, 812 CrossRef CAS PubMed.
- (a) C. Nguyen, J. Grimes, Y. V. Gerasimova and D. M. Kolpashchikov, Chem. – Eur. J., 2011, 17, 13052 CrossRef CAS PubMed; (b) D. M. Kolpashchikov, Scientifica, 2012, 2012, 928783 CrossRef PubMed; (c) D. M. Kolpashchikov, Y. V. Gerasimova and M. S. Khan, ChemBioChem, 2011, 12, 2564 CrossRef CAS PubMed; (d) H. N. Bengtson and D. M. Kolpashchikov, ChemBioChem, 2014, 15, 228 CrossRef CAS PubMed.
- Y. V. Gerasimova and D. M. Kolpashchikov, Chem. Soc. Rev., 2014, 43, 6405–6438 RSC.
- J. Zheng, R. Yang, M. Shi, C. Wu, X. Fang, Y. Li, J. Li and W. Tan, Chem. Soc. Rev., 2015, 44, 3036 RSC.
- A. Tsourkas, M. A. Behlke, S. D. Rose and G. Bao, Nucleic Acids Res., 2003, 31, 1319 CrossRef CAS PubMed.
- J. Sanders Sevall, Mol. Cell. Probes, 2000, 14, 249 CrossRef CAS PubMed.
- A. Emadi, M. T. Crim, D. J. Brotman, A. J. Necochea, L. Samal, L. M. Wilson, E. B. Bass and J. B. Segal, Am. J. Hematol., 2010, 85, 264 CrossRef CAS PubMed.
- (a) K. A. Browne, J. Am. Chem. Soc., 2005, 127, 1989 CrossRef CAS PubMed; (b) Y. Kim, C. J. Yang and W. Tan, Nucleic Acids Res., 2007, 35, 7279 CrossRef CAS PubMed; (c) Y. Ueno, A. Kawamura, K. Takasu, S. Komatsuzaki, T. Kato, S. Kuboe, Y. Kitamura and Y. Kitade, Org. Biomol. Chem., 2009, 7, 2761 RSC; (d) C. Crey-Desbiolles, D. R. Ahn and C. J. Leumann, Nucleic Acids Res., 2005, 33, e77 CrossRef PubMed; (e) P. Sheng, Z. Yang, Y. Kim, Y. Wu, W. Tan and S. A. Benner, Chem. Commun., 2008, 5128 RSC.
- (a) A. Carmon, T. J. Vision, S. E. Mitchell, T. W. Thannhauser, U. Müller and S. Kresovich, BioTechniques, 2002, 32, 410 CAS; (b) K. M. Carneiro, F. A. Aldaye and H. F. Sleiman, J. Am. Chem. Soc., 2010, 132, 679 CrossRef CAS PubMed; (c) P. Chidchob, T. G. Edwardson, C. J. Serpell and H. F. Sleiman, J. Am. Chem. Soc., 2016, 138, 4416 CrossRef CAS PubMed; (d) D. Whitcombe, J. Theaker, S. P. Guy, T. Brown and S. Little, Nat. Biotechnol., 1999, 17, 804 CrossRef CAS PubMed; (e) K. R. Fox, E. Flashman and D. Gowers, Biochemistry, 2000, 39, 6714 CrossRef CAS PubMed.
- D. V. Pyshnyi, A. A. Lomzov, I. A. Pyshnaya and E. M. Ivanova, J. Biomol. Struct. Dyn., 2006, 23, 567 CAS.
- A. Y. Skoblov, M. V. Vichuzhanin, V. M. Farzan, O. A. Veselova, T. A. Konovalova, A. T. Podkolzin, G. A. Shipulin and T. S. Zatsepin, Bioorg. Med. Chem., 2015, 23, 6749 CrossRef CAS PubMed.
- M. Kreutz, G. Schock, J. Kaiser, N. Hochstein and R. Peist, Methods Mol. Biol., 2015, 1315, 17 Search PubMed.
- J. F. Ferguson, C. M. Phillips, J. McMonagle, P. Pérez-Martínez, D. I. Shaw, J. A. Lovegrove, O. Helal, C. Defoort, I. M. Gjelstad, C. A. Drevon, E. E. Blaak, W. H. Saris, I. Leszczyńska-Gołabek, B. Kiec-Wilk, U. Risérus, B. Karlström, J. Lopez-Miranda and H. M. Roche, Atherosclerosis, 2010, 211, 539 CrossRef CAS PubMed.
- K. Juul, A. Tybjaerg-Hansen, P. Schnohr and B. G. Nordestgaard, Ann. Intern. Med., 2004, 140, 330 CrossRef CAS PubMed.
- A. A. Litonju, L. Gong, Q. L. Duan, J. Shin, M. J. Moore, S. T. Weiss, J. A. Johnson, T. E. Klein and R. B. Altman, Pharmacogenet. Genomics, 2010, 20, 64 CrossRef PubMed.
- (a) A. N. Chuvilin, M. V. Serebryakova, I. P. Smirnov and G. E. Pozmogova, Bioconjugate Chem., 2009, 20, 1441 CrossRef CAS PubMed; (b) A. N. Chuvilin, I. P. Smirnov, A. G. Mosina, A. M. Varizhuk and G. E. Pozmogova, PLoS One, 2016, 11, e0166911 Search PubMed.
- J. A. Sanchez, K. E. Pierce, J. E. Rice and L. J. Wangh, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 1933 CrossRef CAS PubMed.
- M. Zuker, Nucleic Acids Res., 2003, 31, 3406 CrossRef CAS PubMed.
- N. E. Watkins and J. Santa Lucia, Nucleic Acids Res., 2005, 33, 6258 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Sequences of primers, HPLC and MS spectra for probes. See DOI: 10.1039/c6an02441f |
| This journal is © The Royal Society of Chemistry 2017 |