The effect of “Jelly” CdTe QD uptake on RAW264.7 monocytes: immune responses and cell fate study†
O.
Gladkovskaya
ab,
A.
Loudon
c,
M.
Nosov
d,
Y. K.
Gun'ko
ce,
G. M.
O'Connor
a and
Y.
Rochev
*bf
aSchool of Physics, National University of Ireland, Galway, Ireland
bNetwork of Excellence for Functional Biomaterials, Galway, Ireland
cCRANN and School of Chemistry, Trinity College Dublin, Ireland
dFarmLab Diagnostics, Emlagh, Elphin, Ireland
eITMO University, 197101 Saint Petersburg, Russia
fSchool of Chemistry, National University of Ireland, Galway, Ireland. E-mail: yury.rochev@nuigalway.ie; Fax: (+353) 91 494 596; Tel: (+353) 91 492 806
First published on 12th October 2015
Abstract
Encapsulation of Quantum Dots (QDs) has become an essential factor which regulates particles cytotoxicity, as well as physical and chemical stability. Negatively charged cellular membranes have a great affinity to nanoparticles with surface molecules carrying positive charge, hence creating perfect conditions for fast and aggressive intracellular penetration. The preference for non-charged outer shells is topical in QD design and various applications. In the current paper we develop gelatination as a prominent coating approach to create neutrally passivated QDs with improved biocompatibility. We have revealed the trends in particle's uptake, accumulation, intracellular localisation and retaining time as well as RAW264.7 monocyte cell fate and immune responses. Also the difference in particle endocytosis kinetics and dynamics has been shown to depend on the QD core size. The intracellular QD content along with cell responses at the population level was quantified by flow cytometry.
Introduction
Quantum dots (QDs) are small semiconducting nanoparticles which are composed of a few hundred atoms that leads to quantum confinement effects, high surface-to-volume ratio and consequently to their exceptional optical sensitivity and reactivity.1–4 The exposure of a high number of core atoms to the surface of the quantum dot frequently results in leakage of ions from the particle core and associated free radical formation. Thus non-coated nanoparticles are not suitable for any biological application due to their low compatibility with physiological medium conditions and irregularities in optical parameters. Several strategies have been applied to reduce the QD cytotoxicity including incorporation in micelles and covering with polymers (TOPO, PEG), proteins (albumin), amino acids and sulphur-containing compounds (TGA).Gelatination has been explored as an effective approach to significantly increase the particle biocompatibility without reducing its quantum yield and fluorescence intensity.5 The surface of “Jelly” CdTe QDs has a mixture of functional groups (e.g. amino, carboxyl, mercapto-groups, etc.) due to the nature of gelatin – it consists of fragmented peptides of dehydrolysed collagen, therefore it doesn't have a regular structure. As a result, gelatinated QDs can be linked to biomolecules (proteins, antibodies, oligonucleotides, drugs, etc.) by multiple paths.6,7
Macrophages serve as antigen-presenting cells (APCs) expressing CD80 and CD86 receptors belonging to the B7 superfamily of genes. These two biomolecules (also known as B7-1 and B7-2) play an important role in T-cell activation by providing co-stimulatory signals. T-cell promotion requires either the presence of T-cell receptors (TCRs) or the ligation of CD28 molecules. However binding of CD152 (or so-called CTLA-4, cytotoxic T-lymphocyte antigen-4) opposes T cell initiation. At first sight it appears that there is no difference between CD80 and CD86 molecules: they are complimentary to the same ligands, expressed by the same cell types and have the same functions. The distinct behaviours of these two proteins affect the T-cell fate. CD86 has a higher dissociation/association ability and shorter activation time, and it preferably binds to CD28 ligands. CD80 has more affinity to CD152 receptors, but it is expressed on macrophages after CD86 triggering. It is intriguing that although a quicker CD86–CD28 interaction results in enhanced T-cell activation, the opposite pair, CD80–CD152, has a higher affinity, hence the amplified silencing effect.8
The RAW264.7 murine macrophage-like cell line has been employed in a number of studies due to its quick doubling time, efficacy in internalizing, comparatively easy activation, good host quality for transfection, and expression of an essential set of inflammatory proteins (IL-6, IL-10 and TNF-α) and surface receptors (CD80 and CD86). These adherent cells have monocyte morphology with the potential to be promoted to macrophages under certain conditions, for example when challenged by lipopolysaccharides (LPSs), or in the presence of mannose containing antigens or TLR. This cell line enables a broad use in in vitro biomaterial trials for investigating all kinds of cell–material interactions including cell covering adhesion, cell growth, cell detachment, mitochondrial and proliferation activity, and immune and mitosis profiling.9,10 Alternatively, the RAW264.7 cell line can be considered as a reasonable first approach for examining the nanoparticle fate when injected into the blood stream, followed by bio-imaging and the final cleavage.
Materials and methods
QD synthesis
CdTe QDs were synthesised according to a previously published procedure.11 Briefly, Al2Te3 reacted with sulphuric acid to produce H2Te gas which was bubbled through an aqueous solution of CdCl2, thioglycolic acid (TGA) and 0.3 g of gelatin, with pH at 11. The molar ratio of Cd![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Te
Te![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TGA was 1
TGA was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.25
0.25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.4. The reaction mixture was then heated under reflux for 2 to 48 hours depending on the desired nanoparticle size. Narrow size distribution fractions were collected via size-selective precipitation using isopropanol.
1.4. The reaction mixture was then heated under reflux for 2 to 48 hours depending on the desired nanoparticle size. Narrow size distribution fractions were collected via size-selective precipitation using isopropanol.
UV-vis and PL spectra
Absorbance was examined on a Shimadzu UV-1601 spectrophotometer; distilled water was taken as the baseline. PL spectra were recorded on a Cary Eclipse spectrometer. All measurements were performed to characterize the optical properties of the nanoparticles obtained.Cell culture
The RAW 264.7 murine macrophage cell line was used in this study. Cells were cultured in Dulbecco's Modified Eagle Media (DMEM; Sigma), supplemented with 10% Fetal Bovine Serum (FBS; Sigma), 100 μg mL−1 of penicillin and 100 μg mL−1 of streptomycin. Macrophages were maintained under a humidified atmosphere with 5% CO2 at 37 °C.Transmission electron microscopy (TEM)
Monocytes were seeded onto thermanox films (13 mm diameter) in a 24 well-plate. The seeding density was 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well. The cells were cultured for 24 hours; after that, conditioned culture medium was replaced by fresh media containing QDs in an appropriate concentration. Cells were further incubated in the presence of nanoparticles for next 12 or 24 hours as desired. Cells were fixed with 2.5% glutaraldehyde and post-fixed with 1% osmium. The fixed samples were introduced into an ethanol gradient and embedded into low viscosity resin (TAAB, Berks, England). The obtained blocks were trimmed and proceeded to fine section cut. Samples were imaged using a Hitachi H7000 transmission electron microscope.
000 cells per well. The cells were cultured for 24 hours; after that, conditioned culture medium was replaced by fresh media containing QDs in an appropriate concentration. Cells were further incubated in the presence of nanoparticles for next 12 or 24 hours as desired. Cells were fixed with 2.5% glutaraldehyde and post-fixed with 1% osmium. The fixed samples were introduced into an ethanol gradient and embedded into low viscosity resin (TAAB, Berks, England). The obtained blocks were trimmed and proceeded to fine section cut. Samples were imaged using a Hitachi H7000 transmission electron microscope.
ds-DNA quantification
A Quant-iT PicoGreen dsDNA Assay Kit was used for a precise counting of the cell number in the probe. The cells were seeded in a 24-well plate to a density of 1 × 105 cells per well, 24 hours prior to the experiment. Different types of QDs (either TGA or TGA-gelatin-covered) within a range of concentrations (1–100 nM final concentration) were added to macrophages. After 24 hours of co-incubation, the cells were subjected to the PicoGreen assay according to the protocol.Annexin V apoptosis assay
In this assay cells were seeded to a density of 2.5 × 105 cells per well in 6 well-plates. After 24 hours of culture, appropriate amounts of QDs were added to each well. Control samples remained untreated. Cells were co-incubated with or without nanoparticles for 12 or 24 hours. Samples were harvested on the day of analysis. Briefly, the reduced medium was removed and the cells were washed twice with phosphate buffered saline (PBS). Macrophages were harvested by pipetting in fresh media and then were placed in Eppendorf tubes. Cells were washed twice with PBS immediately after harvesting, re-suspended in 500 μl buffer and stained with a viability dye according to the protocol. Afterwards cells were washed with serum-containing buffer. Finally, cells were prepared and stained with the Annexin V Apoptosis Assay Kit (eBioscience) and directly subjected to flow cytometry. All measurements were performed on a BD FACS Canto A fitted with 2 lasers (blue, 488 nm; red, 633 nm) and 6 available colours. Unstained cells, single-stained samples, and cells treated with QDs only (without further staining) were used as quality controls.QD uptake and CD80/86 surface marker expression
Flow cytometry was used to detect the amount of internalized nanoparticles and to measure the expression of pro-inflammatory receptors caused by exposure to QDs. All measurements were performed on a BD FACS Canto A. In this experiment cells were seeded into 6-well plates to a density of 2.5 × 105 cells per well and left 24 hours to adhere. The next day, macrophages were loaded with red or green gelatin coated QDs within a range of concentrations (1–100 nM final concentration). After 12 hours (for the CD86 study) and 24 hours of treatment (for the CD80 study), the probes were subjected to the assay according to the standard protocol. Armenian hamster IgG and rat IgG2a K were used as isotype controls for CD80 and CD86, respectively. All antibodies and isotype controls were purchased from BioLegend. The standard staining protocol recommended by the manufacturer was employed. APC and FITC channels were used as references for signal detection. FlowJo software was used for interpretation of results.Quantification of QDs
The amount of the ingested QD nano-crystals was defined by FlowJo software. At least 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were recorded per tube. A consistent macrophage population was selected from the light scatter graph, and the level of fluorescence in the FITC channel was evaluated from a histogram plot; the geometric mean value was used quantitatively as a statistical parameter. The percentage of the population of interest was found from the overlay of two histograms of cells treated with QDs and untreated controls in the reference channel.
000 events were recorded per tube. A consistent macrophage population was selected from the light scatter graph, and the level of fluorescence in the FITC channel was evaluated from a histogram plot; the geometric mean value was used quantitatively as a statistical parameter. The percentage of the population of interest was found from the overlay of two histograms of cells treated with QDs and untreated controls in the reference channel.
Statistical analysis
A two-tailed unpaired t-test has been used to evaluate the statistical significance of the results. The experiments were compared with the control group. The results were recognised as statistically significant if the p-value is less than 0.05; they're marked with the asterisk symbol (*) in the graphics. All the p-values are given in Table 1, ESI.†Results
Physico-chemical properties of QDs
The as-obtained nanoparticles have been fully characterized. Table 1 shows the main properties of QDs. Both batches have a 29 nm Stokes shift and similar negative surface charge.| Sample name | Absorption, nm | Emission, nm | Core size, nm | Zeta-potential, mV | Standard deviation of zeta-potential |
|---|---|---|---|---|---|
| Green gel | 515 | 546 | 2.7 | −61.7 | 2.1 |
| Red gel | 600 | 629 | 3.7 | −52.3 | 1.4 |
ds-DNA quantification by PicoGreen
Only exposure to the highest concentration (100 nM) had affected the cell viability (Fig. 1). The number of cells was reduced to 36–40% compared to untreated cells after 24 hours of co-incubation. It should be noted that the results of the test reflect the number of live cells in the sample on the day of acquisition, regardless of nanoparticle internalisation. The cell doubling time should be added to contributing factors. Exposure to low concentrations (1 and 10 nM) did not affect the cell viability.QD uptake evaluation by flow cytometry
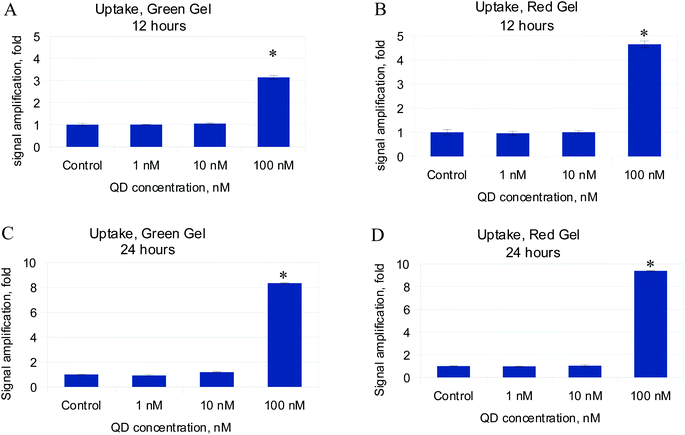
Flow cytometry was employed as a quick and simple acquisition method for nanoparticle ingestion. It allows quantification of the intracellular amount of QDs at the population level by measuring the change in the fluorescence and light scatter pattern in each individual cell. It's been shown that either red or green QDs are accumulating in monocytes over time (Fig. 2); the signal intensity in reference channels has doubled from 12 to 24 hours. The fluorescence response is higher for red QDs. As is observed in the PicoGreen assay, the cell number has not been affected when treated with low doses. We speculate that a threshold should be achieved to promote cellular reaction on the introduced species.12,13CD80/CD86 surface molecule expression
Monocytes are professional phagocytic cells aimed to ingest and destroy foreign bodies or trigger further signaling and consequent T cell activation. Change in CD80/86 surface marker expression evokes inflammatory pathway cascades and activation to macrophages. The marker levels were measured with respect to nanoparticle treatment. Fig. 3 depicts the results of the experiment. Both markers were significantly down-regulated when the cells are introduced to 100 nM concentration regardless of the QD size (5.3 nm for red and 2.3 nm for green). Compared to control cultures, the levels of CD86 were elevated (20–40%) in the case of red QDs (p < 0.005) when treated with 1 and 10 nM.Electron microscopy
The vesicles with trapped QDs are found within the cytoplasm; cells maintain a healthy morphology despite uptake. Fig. 4 shows the obtained TEM images. No obvious hallmarks of apoptosis or necrosis were observed. The nucleus is smooth, and chromatin is not condensed. The only difference between the control and treated cells is the increased number of endosomes. However, the number of cells in the sections is much smaller than that analysed in any other assay. | ||
| Fig. 4 TEM images of untreated monocytes (A), cells treated with 100 nM red (B) and green (C) gelatinated QDs. More images are provided in the ESI.† | ||
Annexin V apoptosis test
The conditioned cell cultures were examined for the prevalent cell fate. The Annexin V detection kit was employed to distinguish live, apoptotic and necrotic stages. The exposure to 1 or 10 nM of nanoparticles did not show any deviations from the control at any time point. The picture has drastically changed when monocytes received 100 nM QDs. The uptake rate did not change for green gelatin coated QDs. The intracellular content of red particles has increased twice from 12 to 24 hours and reached over 90% (Fig. 5A–D). Then, this green or red positive subset was divided into 4 quadrants to quantify viable, early apoptotic, late apoptotic and necrotic cells according to annexin V/viability dye staining (Fig. 5E–H). The majority of cells treated with green QDs remained alive during both control observations. The number of healthy cells also was constant. When the cells were co-cultured with red nanoparticles, they mainly appeared to be dead by the necrosis pathway.Discussion
In our work we used serum-containing culture media to emulate physiological medium conditions. The formation of a protein corona is one of the key events defying further the cell–particle interaction and can't be neglected.14–17 To reveal the potential effects caused by long exposure to QDs, continuous cell culture is required, which is not feasible in a serum-free environment. Wang18 showed that the ingestion pathway, rate and cytotoxicity are not the same once serum proteins are involved. Rapid intake of small amounts of QDs, by cells other than professional phagocytes, has been shown in a number of studies.18–21 Pulse co-incubation (typically up to a few hours) with bare nanoparticles in solution was performed, with an excellent outcome in terms of a high rate of targeting with no or very little cell death. This system is a good first approach for efficacy evaluation. However “real life” cell targeting and drug delivery have more complex routes than the direct cargo–target contact.The drastic difference in the uptake kinetics pattern is exemplified in Fig. 5. For green-emitting nanoparticles the uptake rate hasn't changed from 12 to 24 hours (66 and 67% respectively), or the number of viable cells (93 and 90%). The histogram peak shifts to the right (higher fluorescence), proving QD accumulation over time. As was described by Aberg,22 in continuous exposure to nanoparticles the internalisation is a heterogeneous process and depends on the phase of the cell cycle. According to their study, the intracellular amount of nanoparticles can be ranked as G2/M > S > G1.22,23 Apparently, in long cultures (longer than one cell division cycle) with neutral nanoparticles two processes are competing: accumulation and export. In the case of toxic species a third parameter is contributing, namely cell death and subsequent nanoparticle release to media. According to that, the diagram tail (Fig. 5B) is the signal from cell accumulated QDs (dividing cells) and the rest of the histogram represents the average response from cells in the S/G1 phase. A small amount of cells goes through the apoptotic (5.2 and 7.2% at 12 and 24 hours) or necrotic pathway (1.7 and 2.2% respectively).
The uptake heterogeneity concept is in striking agreement with the results of the experiment involving red-emitting nanoparticles. If we assume that active QD phagocytosis takes place only in one phase of cell cycle, then, after first 12 hours of QD exposure, we will have a cellular subset with significantly high intracellular content of nanoparticles. Considering that cytotoxicity is a cumulative parameter, this subset is likely to die. In next 12 hours necrotic cells are eliminated from the system. The remaining cells proceed through the proliferation cycle again and accumulate more QDs. It results in a strong fluorescence intensity peak shift to the right and an increase in the number of dead cells.
The QD size contributes to the uptake dynamics. Chithrani and Chan24,25 have found the preferred QD size for efficient ingestion. This has been explained by the dependence of the wrapping time on the diameter of primary vesicles when loaded with nanoparticles. According to the study, the optimal diameter for spherical particles is 50 nm. This result was confirmed by Osaki.26 Nevertheless, when the core size of QDs does not exceed 10 nm, the protein corona increases the hydrodynamic size up to hundreds of nanometers. Similarly, Wang18 and Jiang27 suggested that if only large clusters of nanoparticles are formed locally, ingestion might occur. Apart from that, the mechanism and parameters defying uptake are still under discussion. It has been agreed that uptake is an energy dependent process for particles with a core size of 5 nm and above; smaller dots can be transported passively.18 Red-emitting QDs enter the cells via the clathrin-mediated route,21,27,28 however Zhang and Monteiro-Riviere20 have found the caveolae/lipid raft as the endocytosis mechanism via the G protein receptor pathway and the low-density lipoprotein (LDL)/scavenger receptor. Also there's no solidarity in the questions whether the surface coating/charge20,21 influences the uptake or makes no difference.28,29 And whether it is more important than the hydrodynamic size or not.30 The observed contradictions may be related to different cell types used in the experiments.
Conclusion
In the current study we investigated the behaviour of gelatin coated QDs under serum-containing conditions and their interaction with the cells in continuous cultures. Following earlier research, suggesting distinct patterns from that in protein-free media,18 elevation of the toxic dose (100 nM over 10 nM in previous studies) and different cellular responses to the exposed dots of various sizes (2.7 and 3.7 nm) for a time greater than the cell cycle were confirmed. Our results suggest that heterogeneity in the pace of uptake depends on the cell cycle phase. Unlike pulse treatment, where QDs were co-incubated for a short time and particles were captured regardless of the cell cycle phase, in our study it's one of the contributing parameters in endocytosis kinetics.Only 100 nM concentration is considered to affect the cell function. Surprisingly, surface marker expression levels have dropped down to less than 50% from the control. Either green or red QDs drastically decrease the cell number at 100 nM concentration. In the case of red QDs massive cell death via necrosis was observed; this occurred with twice the uptake rate at the 24 hours acquisition point (from 50 to almost 100%). Overall, both QD types tend towards an intracellular occupancy and have a longer retention time when compared with less passivated particles. The ingested nanoparticles form conglomerates and are trapped into the endosomes, clearly observable in the cytoplasm (Fig. 4).
Funding sources
This work was conducted under the framework of INSPIRE, the Irish Government's Programme for Research in Third Level Institutions Cycle 5, National Development Plan 2007–2013 with the assistance of the European Regional Development Fund, the Science Foundation Ireland (SFI 12/IA/1300 project) and the Ministry of Education and Science of the Russian Federation (Grant No. 14.B25.31.0002).Conflicts of interests
None declared.Acknowledgements
The authors are grateful to Shirley Hanley (PhD, NCBES) for help with flow cytometry experiments and Pierce Lalor (Anatomy Department, NUIG) for support with TEM processing and imaging.References
- D. Bera, L. Qian, T.-K. Tseng and P. H. Holloway, Quantum Dots and Their Multimodal Applications: A Review, Materials, 2010, 3, 2260–2345 CrossRef CAS PubMed.
- Y. Zhang and A. Clapp, Overview Of Stabilizing Ligands For Biocompatible Quantum Dot Nanocrystals, Sensors, 2011, 11, 11036–11055 CrossRef PubMed.
- U. Resch-Genger, M. Grabolle, S. Cavaliere-Jaricot, R. Nitschke and T. Nann, Quantum Dots Versus Organic Dyes As Fluorescent Labels, Nat. Methods, 2008, 5(9), 763–775 CrossRef CAS PubMed.
- A. P. Alivisatos, W. Gu and C. Larabell, Quantum Dots As Cellular Probes, Annu. Rev. Biomed. Eng., 2005, 7, 55–76 CrossRef CAS PubMed.
- B. R. Prasad, G. Mullins, N. Nikolskaya, D. Connolly, T. J. Smith, V. A. Gerard, S. J. Byrne, G. L. Davies, Y. K. Gun'ko and Y. Rochev, Effects of long-term exposure of gelatinated and non-gelatinated cadmium telluride quantum dots on differentiated PC12 cells, J. Nanobiotechnol., 2012, 10, 1–14 CrossRef PubMed , Article Number: 4.
- V. A. Gérard, C. M. Maguire, D. Bazou and Y. K. Gun'ko, Folic acid modified gelatin coated quantum dots as potential reagents for in vitro cancer diagnostics, J. Nanobiotechnol., 2011, 9, 50 CrossRef PubMed.
- E. Jan, S. J. Byrne, M. Cuddihy, A. M. Davies, Y. Volkov, Y. K. Gun'ko and N. A. Kotov, High Content Screening as a Universal Tool for Fingerprinting of Cytotoxicity of Nanoparticles, ACS Nano, 2008, 2(5), 928–938 CrossRef CAS PubMed.
- D. M. Sansom, C. N. Manzotti and Y. Zheng, What's the difference between CD80 and CD86?, Trends Immunol., 2003, 24(6), 313–318 CrossRef.
- K. Unfried, C. Albrecht, L.-O. Klotz, A. Von Mikecz, S. Grether-Beck and R. P. F. Schins, Cellular responses to nanoparticles: Target structures and mechanisms, Nanotoxicology, 2007, 1(1), 52–71 CrossRef CAS PubMed.
- M. Hashimoto, H. Toshima, T. Yonezawa, K. Kawai, T. Narushima, M. Kaga and K. Endo, Responses of RAW264.7 macrophages to water-dispersible gold and silver nanoparticles stabilized by metal–carbon r-bonds, J. Biomed. Mater. Res., Part A, 2013, 1–12 Search PubMed.
- S. J. Byrne, Y. Williams, A. Davies, S. A. Corr, A. Rakovich, Y. K. Gunko, Y. P. Rakovich, J. F. Donegan and Y. Volkov, “Jelly Dots”: Synthesis and Cytotoxicity Studies of CdTe Quantum Dot–Gelatin Nanocomposites, Small, 2007, 7, 1152–1156 CrossRef PubMed.
- M. M. Malingré, J. H. Beijnen, H. Rosing, F. J. Koopman, R. C. Jewell, E. M. Paul, W. W. Ten Bokkel Huinink and J. H. M. Schellens, Co-administration of GF120918 significantly increases the systemic exposure to oral paclitaxel in cancer patients, Br. J. Cancer, 2001, 84(1), 42–47 CrossRef PubMed.
- H. Minami, Y. Sasaki, N. Saijo, T. Ohtsu, H. Fujii, T. Igarashi and K. Itoh, Indirect-response model for the time course of leukopenia with anticancer drugs, Clin. Pharmacol. Ther., 1998, 511–520 CrossRef CAS.
- A. Lesniak, F. Fenaroli, M. P. Monopoli, C. Aberg, K. A. Dawson and A. Salvati, Effects of the Presence or Absence of a Protein Corona on Silica Nanoparticle Uptake and Impact on Cells, ACS Nano, 2012, 6(7), 5845–5857 CrossRef CAS PubMed.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch, F. Baldelli Bombelli and K. A. Dawson, Physical-Chemical Aspects of Protein Corona: Relevance to in Vitro and in Vivo Biological Impacts of Nanoparticles, J. Am. Chem. Soc., 2011, 133, 2525–2534 CrossRef CAS PubMed.
- A. Lesniak, A. Campbell, M. P. Monopoli, I. Lynch, A. Salvati and K. A. Dawson, Serum heat inactivation affects protein corona composition and nanoparticle uptake, Biomaterials, 2010, 31, 9511–9518 CrossRef CAS PubMed.
- S. Tenzer, D. Docter, J. Kuharev, A. Musyanovych, V. Fetz, R. Hecht, F. Schlenk, D. Fischer, K. Kiouptsi, C. Reinhardt, K. Landfester, H. Schild, M. Maskos, S. K. Knauer and R. H. Stauber, Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology, Nat. Nanotechnol., 2013, 8, 772–781 CrossRef CAS PubMed.
- T. Wang and X. Jiang, Size-Dependent Stability of Water-Solubilized CdTe Quantum Dots and Their Uptake Mechanism by Live HeLa Cells, ACS Appl. Mater. Interfaces, 2013, 5, 1190–1196 CAS.
- S. T. Stern, B. S. Zolnik, C. B. McLeland, J. Clogston, J. Zheng and S. E. McNeil, Induction of autophagy in porcine kidney cells by quantum dots: a common cellular response to nanomaterials?, Toxicol. Sci., 2008, 106(1), 140–152 CrossRef CAS PubMed.
- L. W. Zhang and N. A. Monteiro-Riviere, Mechanisms of Quantum Dot Nanoparticle Cellular Uptake, Toxicol. Sci., 2009, 110(1), 138–155 CrossRef CAS PubMed.
- Y. Xiao, S. Pforry, X. Gao, D. R. Holbrook, W. G. Telford and A. Tona, Dynamics and mechanisms of quantum dot nanoparticle cellular uptake, J. Nanobiotechnol., 2010, 8, 13 CrossRef PubMed.
- C. Aberg, J. A. Kim, A. Salvati and K. A. Dawson, Theoretical framework for nanoparticle uptake and accumulation kinetics in dividing cell populations, EPL, 2013, 101, 38007 CrossRef.
- J. A. Kim, C. Åberg, A. Salvati and K. A. Dawson, Role of cell cycle on the cellular uptake and dilution of nanoparticles in a cell population, Nat. Nanotechnol., 2012, 7, 62–68 CrossRef CAS PubMed.
- D. B. Chithrani and W. C. Chan, Elucidating the mechanism of cellular uptake and removal of protein-coated gold nanoparticles of different sizes and shapes, Nano Lett., 2007, 7(6), 1542–1550 CrossRef PubMed.
- B. D. Chithrani, A. A. Ghazani and W. C. W. Chan, Determining the Size and Shape Dependence of Gold Nanoparticle Uptake into Mammalian Cells, Nano Lett., 2006, 6(4), 662–668 CrossRef CAS PubMed.
- F. Osaki, T. Kanamori, S. Sando, T. Sera and Y. Aoyama, A Quantum Dot Conjugated Sugar Ball and Its Cellular Uptake. On the Size Effects of Endocytosis in the Subviral Region, J. Am. Chem. Soc., 2004, 126, 6520–6521 CrossRef CAS PubMed.
- X. Jiang, C. Rocker, M. Hafner, S. Brandholt, R. M. Dorlich and G. U. Nienhaus, Endo- and Exocytosis of Zwitterionic Quantum Dot Nanoparticles by Live HeLa Cells, ACS Nano, 2010, 4(11), 6787–6797 CrossRef CAS PubMed.
- S. S. Minami, B. Sun, K. Popat, T. Kauppinen, M. Pleiss, Y. Zhou, M. E. Ward, P. Floreancig, L. Mucke, T. Desai and L. Gan, Selective targeting of microglia by quantum dots, J. Neuroinflammation, 2012, 9–22 Search PubMed.
- J. P. Ryman-Rasmussen, J. E. Riviere and N. A. Monteiro-Riviere, Surface Coatings Determine Cytotoxicity and Irritation Potential of Quantum Dot Nanoparticles in Epidermal Keratinocytes, J. Invest. Dermatol., 2006, 1–11 Search PubMed.
- C. Dong and J. Irudayaraj, Hydrodynamic Size-Dependent Cellular Uptake of Aqueous QDs Probed by Fluorescence Correlation Spectroscopy, J. Phys. Chem. B, 2012, 116, 12125–12132 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5tx00153f |
| This journal is © The Royal Society of Chemistry 2016 |