Open Access Article
Open Access ArticleDelivery of an active lysosomal enzyme using GNeosomes†
Kristina M.
Hamill
a,
Ezequiel
Wexselblatt
a,
Wenyong
Tong
b,
Jeffrey D.
Esko
b and
Yitzhak
Tor
*a
aDepartment of Chemistry and Biochemistry, University of California, San Diego, La Jolla, CA 92093-0358, USA. E-mail: ytor@ucsd.edu
bCellular and Molecular Medicine, University of California, San Diego, La Jolla, CA 92093-0687, USA
First published on 9th August 2016
Abstract
Two methods for assembling guanidinoneomycin-decorated liposomes are presented and their ability to deliver an active enzyme to the lysosomes and restore enzyme function in diseased cells is compared.
Lysosomes are critical for the degradation of intra- and extracellular material through the action of over 50 acid hydrolases and membrane proteins.1–3 The absence or low activity of a particular lysosomal hydrolase leads to accumulation of its substrate(s) which causes damage in various tissues, organs, and in some cases, the central nervous system. More than 50 recessively inherited lysosomal storage disorders (LSDs) are known. While individually rare, their combined prevalence is about 1 in 8000 births.4–6
The predominant treatment for LSDs is enzyme replacement therapy (ERT), where intravenously administered enzyme is taken up by cells through a mannose-6-phosphate mediated pathway.6–9 Although ERT has been successful in treating several LSDs, it is not equally effective for all enzymes and disorders, and delivery to cartilage, heart valve, skeletal muscle, and the brain is very limited.7–9 In an attempt to improve ERT, lysosomal enzymes were first encapsulated in liposomes 45 years ago.10,11 More recent efforts have focused on attaching targeting ligands, such as low molecular weight ligands (e.g., rhodamine B and mannose-6-phosphate) or high molecular weight proteins (e.g., transferrin), to the surface of liposomes to improve their lysosomal delivery.12–17
We recently reported the assembly and cellular uptake of GNeosomes, lipid vesicles decorated with stearyl-GNeo, an amphiphilic derivative of guanidinoneomycin (GNeo). Although other guanidinium-rich transporters18–23 have been used to improve the intracellular delivery of liposomes,24–31 GNeo is unique in being a highly lysosomotropic transporter capable of delivery through heparan sulfate exclusive pathways.15 In addition to the universal benefits of liposomal packaging, incorporation of GNeo significantly increases the uptake and lysosomal delivery of diverse cargo compared to unmodified liposomes.15
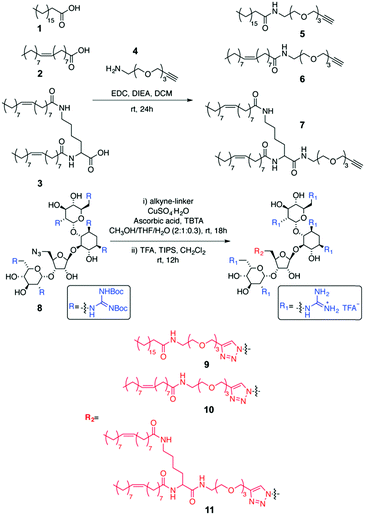
In the above-mentioned approach to assembling GNeosomes, an amphiphilic GNeo derivative (stearyl-GNeo) was introduced during the formation of liposomes. Here, we first report the synthesis of two novel GNeo–lipid derivatives (Scheme 1). We then compare different methods for incorporating the transporter into liposomes (Fig. 1). Advantageous post-insertion and post-modification methods introduce GNeo into pre-formed liposomes thus modifying only their outer surface, without ever premixing the cargo and carrier, thus facilitating the encapsulation of both positively- and negatively-charged cargo. Liposomes with GNeo post-inserted showed enhanced cellular uptake of a small molecule dye compared to the unmodified liposomes and their ability to deliver α-L-iduronidase, a lysosomal enzyme. GNeosomes increased the overall uptake of the enzyme compared to plain liposomes. A sufficient amount of enzyme was delivered to restore the normal turnover of glycosaminoglycans in patient MPS I cells, which lack endogenous enzyme. We conclude that GNeosomes can potentially be used to deliver therapeutic amounts of active enzyme to the lysosomes for the treatment of lysosomal storage disorders.
Three GNeo–lipid derivatives were synthesized from stearic acid (1), oleic acid (2), and a dimeric oleic acid tail (3) as outlined in Scheme 1. Briefly, the fatty acid (1, 2 or 3) was coupled to the amino group of an amino-alkyne-functionalized triethylene glycol (4).32 The resulting compounds (5, 6 and 7) underwent a 1,3-dipolar cycloaddition with 833 followed by acidic deprotection of the Boc-guanidinium groups to yield stearyl-GNeo (9),15 oleyl-GNeo (10, Scheme S1, ESI†), and di-oleyl-GNeo (11, Scheme S2, ESI†). In addition to the GNeo–lipids that can be directly incorporated in the liposomal bilayer, GNeo–NHS (14) was synthesized by clicking a previously reported alkyne-BocGNeo derivative (12)34 to an azide-NHS-functionalized triethylene glycol linker (13, Scheme S3, ESI†) to evaluate post-modification of liposomes (Scheme 2 and Scheme S3, ESI†).
Liposomes were prepared by thin-film hydration followed by freezing and thawing cycles and extrusion. The GNeo–lipids were incorporated into liposomes by either “pre-insertion” or “post-insertion” as described in the ESI† and schematically represented in Fig. 1. Alternatively, the primary amines on the surface of preformed liposomes containing 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) were modified with GNeo–NHS (14, Scheme 2) (Fig. 1, “post-modified liposomes”).
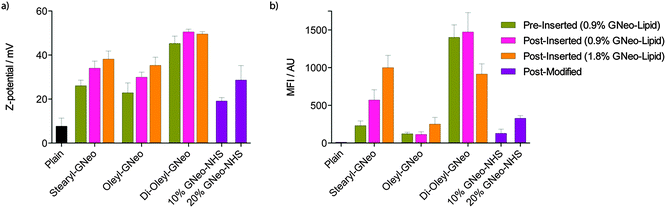
The presence of GNeo on the outer surface of liposomes was confirmed by measuring their zeta potentials. GNeosomes exhibited a positive increase in zeta potential compared to plain liposomes. The zeta potentials of the pre-inserted GNeosomes are lower than the corresponding post-inserted GNeosomes, possibly due to partitioning of GNeo–lipid into both leaflets of the liposomal membrane. Addition of a higher concentration of GNeo–lipid or GNeo–NHS generally resulted in an increase in zeta potential (Fig. 2a). Post-modified liposomes had the lowest zeta potential, but also showed a dependence on the degree of derivatization.
To first evaluate the cellular uptake of the GNeosomes generated by the different preparation methods outlined above, a fluorescent cyanine dye, Cy5, was encapsulated. Uptake was evaluated in wild-type Chinese hamster ovary (CHO-K1) cells and analyzed by flow cytometry. As shown in Fig. 2b, the mean fluorescence intensity (MFI) of cells treated with GNeosomes is significantly higher than cells treated with plain liposomes. Generally, di-oleyl-GNeosomes exhibited the highest uptake, followed by stearyl-GNeosomes. This is consistent with the trend seen for their zeta potentials, and taken together, suggests these lipids insert better into the liposomal membrane, leading to a higher concentration of GNeo on the surface and higher uptake. Oleyl-GNeosomes and GNeo–NHS modified GNeosomes had the lowest zeta potentials and also exhibited the lowest cellular uptake suggesting a lower degree of GNeo modification. The decrease in uptake when a higher concentration of di-oleyl-GNeo was used is attributed to a lower dye encapsulation efficiency (EE), compared to all other preparations (Fig. S1, ESI†). This suggests that the higher concentration of di-oleyl-GNeo might result in leakage of the dye from the liposomes.
To investigate whether GNeosomes can deliver an active enzyme to the lysosomes, α-L-iduronidase (IDUA) was encapsulated. IDUA is a lysosomal enzyme responsible for hydrolyzing the terminal α-L-iduronic acid residues in heparan sulfate (HS) and dermatan sulfate (DS). A deficiency in IDUA leads to the accumulation of HS and DS in the lysosomes and is responsible for the lysosomal storage disease mucopolysaccharidosis I (MPS I; Hurler, Hurler-Scheie, and Scheie syndromes).35
Liposomes post inserted with GNeo on their surface were prepared as described above, replacing the low MW dye with IDUA. The amount of encapsulated enzyme was evaluated by SDS-PAGE (Fig. S2, ESI†) and checked for activity by measuring the conversion of 4-methylumbelliferyl α-L-iduronide into the fluorophore 4-methylumbelliferone (4-MU).36 Cellular uptake was assessed in IDUA-deficient MPS I fibroblasts. The cells were incubated with the liposomes for 1 h at 37 °C then lysed and analyzed for IDUA activity using the above mentioned fluorescence-based assay. Low enzyme activity was observed in cells treated with plain liposomes; on the other hand, cells treated with GNeosomes show more than ten-fold higher enzyme activity than untreated MPS I cells (Fig. 3a). Liposomes post-inserted with 1.8% stearyl-GNeo delivered almost twice as much active enzyme to cells than liposomes post-inserted with 0.9% stearyl-GNeo, similar to the enhanced uptake of Cy5 at higher concentrations of stearyl-GNeo (Fig. 2). The di-oleyl-GNeosomes also exhibited an uptake pattern similar to the delivery of Cy5 with the pre-inserted and post-inserted liposomes behaving similarly.
To determine whether GNeosomes were delivering active IDUA to the lysosomes, a label-chase experiment was performed. MPS I and control fibroblast (HFF) cells were incubated with [35S]-sulfate for 48 h to radiolabel sulfated glycosaminoglycans. The cells were then incubated with plain liposomes or GNeosomes for 1 h at 37 °C, washed, incubated with fresh medium for another 24 h, and analyzed for the amount of [35S]glycosaminoglycans associated with the cells. As shown in Fig. 3b, MPS I fibroblasts store [35S]glycosaminoglycans, whereas control HFF cells turnover glycosaminoglycans (GAGs). Plain liposomes lowered the amount of stored GAGs by about 50%, whereas GNeosomes returned the turnover of GAGs to a level comparable to that found in control HFF cells (Fig. 3b). These results indicate that GNeosomes are therefore taken up by IDUA-deficient fibroblasts, reach the lysosomal compartment, release their cargo and restore IDUA activity.
Uptake was also compared to the GNeo-conjugated enzyme (Fig. 3).37 GNeo–IDUA has previously been shown to have enhanced uptake and activity compared to Aldurazyme, the high-uptake form currently in clinical use for treatment of MPS I patients.37 Maintaining this high level of uptake by GNeo, while encapsulating the enzyme in liposomes, could have additional benefits for in vivo applications, including improved stability typically seen with liposomal delivery systems.38–41
In conclusion, we have demonstrated that introduction of GNeo to the surface of liposomes results in superior uptake of a small molecule dye in wild-type CHO cells compared to unmodified liposomes. These GNeosomes were also demonstrated to deliver and release an active enzyme to the lysosomes in MPS I human fibroblasts. The HS selectivity of GNeo and its efficacious lysosomal delivery results in a unique delivery system. Because virtually all mammalian cells express heparan sulfate, GNeosomes could be ideal for improving the enzymatic treatment of lysosomal storage disorders that affect all tissues. Furthermore, lipid vesicles can be used to entrap other lysosomal enzymes whose activity might be affected by direct conjugation.
Acknowledgements
This work was supported by grant GM077471 and a grant from the MPS Society. We acknowledge the UCSD Chemistry and Biochemistry Mass Spectrometry Facility and the UCSD Inorganic Materials Characterization Facility.Notes and references
- J. P. Luzio, P. R. Pryor and N. A. Bright, Nat. Rev. Mol. Cell Biol., 2007, 8, 622–632 CrossRef CAS PubMed.
- T. Braulke and J. S. Bonifacino, Biochim. Biophys. Acta, Mol. Cell Res., 2009, 1793, 605–614 CrossRef CAS PubMed.
- H. Appelqvist, P. Waster, K. Kagedal and K. Ollinger, J. Mol. Cell Biol., 2013, 5, 214–226 CrossRef CAS PubMed.
- E. F. Neufeld, Annu. Rev. Biochem., 1991, 60, 257–280 CrossRef CAS PubMed.
- B. Winchester, A. Vellodi and E. Young, Biochem. Soc. Trans., 2000, 28, 150–154 CrossRef CAS PubMed.
- C. W. Richard III, in Introduction to Biological and Small Molecule Drug Research and Development, ed. S. R. Jefferis, Elsevier, Oxford, 2013, pp. 327–341 Search PubMed.
- G. A. Grabowski and R. J. Hopkin, Annu. Rev. Genomics Hum. Genet., 2003, 4, 403–436 CrossRef CAS PubMed.
- M. Beck, IUBMB Life, 2010, 62, 33–40 CAS.
- L. Martin-Banderas, M. A. Holgado, M. Duran-Lobato, J. J. Infante, J. Alvarez-Fuentes and M. Fernandez-Arevalo, Curr. Med. Chem., 2016, 23, 929–952 CrossRef CAS PubMed.
- G. Gregoriadis and B. E. Ryman, Eur. J. Biochem., 1972, 24, 485–491 CrossRef CAS PubMed.
- G. Gregoriadis and B. E. Ryman, Biochem. J., 1972, 129, 123–133 CrossRef CAS PubMed.
- A. Koshkaryev, R. Thekkedath, C. Pagano, I. Meerovich and V. P. Torchilin, J. Drug Targeting, 2011, 19, 606–614 CrossRef CAS PubMed.
- R. Thekkedath, A. Koshkaryev and V. P. Torchilin, Nanomedicine, 2013, 8, 1055–1065 CrossRef CAS PubMed.
- E. Crucianelli, P. Bruni, A. Frontini, L. Massaccesi, M. Pisani, A. Smorlesi and G. Mobbili, RSC Adv., 2014, 4, 58204–58207 RSC.
- E. Wexselblatt, J. D. Esko and Y. Tor, ACS Nano, 2015, 9, 3961–3968 CrossRef CAS PubMed.
- A. Koshkaryev, A. Piroyan and V. P. Torchilin, Cancer Biol. Ther., 2012, 13, 50–60 CrossRef CAS PubMed.
- S. Muro, E. H. Schuchman and V. R. Muzykantov, Mol. Ther., 2006, 13, 135–141 CrossRef CAS PubMed.
- E. Vives, P. Brodin and B. Lebleu, J. Biol. Chem., 1997, 272, 16010–16017 CrossRef CAS PubMed.
- S. Fawell, J. Seery, Y. Daikh, C. Moore, L. L. Chen, B. Pepinsky and J. Barsoum, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 664–668 CrossRef CAS.
- P. A. Wender, D. J. Mitchell, K. Pattabiraman, E. T. Pelkey, L. Steinman and J. B. Rothbard, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 13003–13008 CrossRef CAS PubMed.
- C. Bechara and S. Sagan, FEBS Lett., 2013, 587, 1693–1702 CrossRef CAS PubMed.
- E. G. Stanzl, B. M. Trantow, J. R. Vargas and P. A. Wender, Acc. Chem. Res., 2013, 46, 2944–2954 CrossRef CAS PubMed.
- E. Wexselblatt, J. D. Esko and Y. Tor, J. Org. Chem., 2014, 79, 6766–6774 CrossRef CAS PubMed.
- V. P. Torchilin, Biopolymers, 2008, 90, 604–610 CrossRef CAS PubMed.
- V. P. Torchilin, R. Rammohan, V. Weissig and T. S. Levchenko, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 8786–8791 CrossRef CAS PubMed.
- C. Marty, C. Meylan, H. Schott, K. Ballmer-Hofer and R. A. Schwendener, Cell. Mol. Life Sci., 2004, 61, 1785–1794 CAS.
- Y. L. Tseng, J. J. Liu and R. L. Hong, Mol. Pharmacol., 2002, 62, 864–872 CrossRef CAS PubMed.
- M. M. Fretz, G. A. Koning, E. Mastrobattista, W. Jiskoot and G. Storm, Biochim. Biophys. Acta, Biomembr., 2004, 1665, 48–56 CrossRef CAS PubMed.
- M. Furuhata, H. Kawakami, K. Toma, Y. Hattori and Y. Maitani, Bioconjugate Chem., 2006, 17, 935–942 CrossRef CAS PubMed.
- S.-A. Cryan, M. Devocelle, P. J. Moran, A. J. Hickey and J. G. Kelly, Mol. Pharmaceutics, 2006, 3, 104–112 CrossRef CAS PubMed.
- B. Chatin, M. Mevel, J. Devalliere, L. Dallet, T. Haudebourg, P. Peuziat, T. Colombani, M. Berchel, O. Lambert, A. Edelman and B. Pitard, Mol. Ther. – Nucleic Acids, 2015, 4, e244 CrossRef CAS PubMed.
- A. Natarajan, W. Du, C. Y. Xiong, G. L. DeNardo, S. J. DeNardo and J. Gervay-Hague, Chem. Commun., 2007, 695–697 RSC.
- J. L. Childs-Disney, M. Wu, A. Pushechnikov, O. Aminova and M. D. Disney, ACS Chem. Biol., 2007, 2, 745–754 CrossRef CAS PubMed.
- A. V. Dix, L. Fischer, S. Sarrazin, C. P. H. Redgate, J. D. Esko and Y. Tor, ChemBioChem, 2010, 11, 2302–2310 CrossRef CAS PubMed.
- G. Bach, R. Friedman, B. Weissmann and E. F. Neufeld, Proc. Natl. Acad. Sci. U. S. A., 1972, 69, 2048–2051 CrossRef CAS.
- J. J. Hopwood, V. Muller, A. Smithson and N. Baggett, Clin. Chim. Acta, 1979, 92, 257–265 CrossRef CAS.
- S. Sarrazin, B. Wilson, W. S. Sly, Y. Tor and J. D. Esko, Mol. Ther., 2010, 18, 1268–1274 CrossRef CAS PubMed.
- E. Mayhew, D. Papahadjopoulos, Y. M. Rustum and C. Dave, Cancer Res., 1976, 36, 4406–4411 CAS.
- H.-I. Chang and M.-K. Yeh, Int. J. Nanomed., 2012, 7, 49–60 CAS.
- T. M. Allen and P. R. Cullis, Adv. Drug Delivery Rev., 2013, 65, 36–48 CrossRef CAS PubMed.
- B. S. Pattni, V. V. Chupin and V. P. Torchilin, Chem. Rev., 2015, 115, 10938–10966 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials, synthesis and characterization of new compounds, preparation and characterization of liposomes, and encapsulation efficiencies. See DOI: 10.1039/c6tb01387b |
| This journal is © The Royal Society of Chemistry 2016 |