Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synthesis of diblock/statistical cationic glycopolymers with pendant galactose and lysine moieties: gene delivery application and intracellular behaviors†
Jingjing
Sun
a,
Ruilong
Sheng
*ab,
Ting
Luo
a,
Zhao
Wang
a,
Hui
Li
a and
Amin
Cao
*a
aKey Laboratory of Synthetic and Self-assembly Chemistry for Organic Functional Molecules, Shanghai Institute of Organic Chemistry, CAS. Lingling Road 345, Shanghai, 200032, China. E-mail: rlsheng@mail.sioc.ac.cn; acao@mail.sioc.ac.cn; Tel: +86-21-54925535
bDepartment of Chemistry, Université de Montréal, Succursale Centre-ville, Montreal, Quebec H3C3J7, Canada
First published on 9th June 2016
Abstract
A new series of cationic block copolymers PHML-b-PMAGal and statistical copolymers P(HML-st-MAGal) with pendant natural galactose and (L-)-lysine moieties were prepared via RAFT (reversible addition–fragmentation chain-transfer) polymerization. The block/statistical copolymers showed a high plasmid DNA binding affinity (N/P < 2) and the as-formed polyplexes were spherical nanoparticles with the average size of 100–300 nm and surface zeta potentials of +30.2 to +46.3 mV. The cytotoxicity and gene transfection efficacy of the PHML-b-PMAGal and P(HML-st-MAGal) vectors strongly depend on the polymer architectures (block/statistical) and the galactose content. Notably, the statistical copolymer P(HML40-st-MAGal4) with 4.8% galactose content showed the highest gene transfection efficiency among the synthesized cationic polymers, 6.8-fold higher than that of the “gold standard” bPEI-25k in the presence of 10% FBS (fetal bovine serum) in various cell lines. An intracellular uptake mechanism (with 10% FBS) study demonstrated that the P(HML40-st-MAGal4)/pDNA polyplexes entered H1299 cells mainly through caveolae-mediated endocytosis and microtubule-dependent endocytosis pathways. Moreover, the fluorescence imaging study showed that the P(HML40-st-MAGal4)/pDNA polyplexes possessed an obvious “lysosomal escaping” effect that led to efficient pDNA release, which might interpret the fact of the significant increase of the related gene transfection efficiency. Moreover, it could be anticipated that the P(HML40-st-MAGal4) cationic glycopolymer might be employed as a low toxic, highly efficient and serum-compatible gene carrier for practical applications.
1. Introduction
In the past few decades, cationic polymeric non-viral gene vectors have been developed as a new technology for gene therapy, and it is well known that successful gene therapy largely depends on the choice of suitable building blocks in the construction of the cationic polymeric gene vectors.1–4 Although many cationic polymeric vectors were developed to mediate efficient intracellular gene transfection, some defects such as high cytotoxicity,5 immunogenicity6 and low serum-compatibility7,8 severely restricted their clinical applications in gene therapy. To solve these issues, a general approach adopted in previous works was conjugation/modification of cationic polymers with polyethylene glycol (PEG), which could be employed to shield cationic surface charges, decrease cytotoxicity and improve serum-compatibility of cationic polymer vector/gene complexes.5,9 However, it should be noted that the “stealth” effect of PEG could decrease the cell adhesion and the intracellular uptake, further resulting in a lower transfection efficiency of the polyplexes.10 Moreover, the PEG-modified polymeric vectors might induce immune-response as antigens and would be removed by macrophage systems after injection.11Recently, several biocompatible building blocks such as zwitterionic molecules12,13 and carbohydrates14 have been developed as alternative substitutes to PEG moieties in gene/drug delivery systems. Among them, carbohydrates are highly biocompatible natural products with good hydrophilicity and biodegradable characteristics. It had been disclosed that cationic glycopolymer vectors constructed by carbohydrates and cationic units exhibited low cytotoxicity and serum-resistance, which resembled that of the PEG-conjugated polycations due to the positive charge shielding effect. But unlike PEG, the carbohydrate units in glycopolymers have the advantage of enhancing pDNA binding affinity through hydrogen bond interactions, which may further facilitate high gene transfection efficiency.15–18 Besides, the multi-hydroxyl groups on carbohydrate units offered them the possibility for further functional derivation/modification.19,20 For example, Azzam et al.21 synthesized some polyethylenimine and spermine modified dextran and branched-starch cationic polymers, and found that the gene transfection capability largely relied upon the polysaccharide structures, and dextran-derived cationic polymers showed the highest transfection capability. Davis et al.22 reported that the length of cationic blocks played essential roles in the cytotoxicity and gene transfection of cyclodextrin-based cationic gene vectors. Gu et al.23 prepared (L)-lysine-modified chitosan polymers, which exhibited high in vitro gene transfection capability and low cytotoxicity. However, due to the complicated and uncontrollable structures of these natural polysaccharide glycopolymers, it is difficult to elucidate their concise structure–activity relationships. Thus, developing new cationic glycopolymers with defined and controllable molecular structures as efficient gene vectors/carriers is of high demand.
With the development of controlled free radical polymerization, a lot of functional glycopolymers with well-defined structures and controllable molecular-weight could be facilely prepared,24,25 which made it possible for further investigating the correlation between polymer structure and gene transfection related properties. For example, Narain et al. synthesized a series of well-defined block and random glycopolymers as gene vectors and found that the latter showed a higher gene transfection efficiency than the former one.26–29 Reineke et al. prepared some cationic glycopolymers as gene vectors and disclosed that introducing sugar-containing blocks could increase the stability of pDNA-loaded complexes by preventing their aggregation in culture medium.14,30–33 However, to our knowledge, many cationic building blocks used for constructing glycopolymeric gene vectors were non-biodegradable or less-biocompatible. Alternatively, cationic amino acids (such as (L)-lysine and (L)-arginine) serve as essential biocompatible natural cationic building blocks for constructing new cationic gene vectors with low cytotoxicity and high pDNA binding affinity.34,35 Moreover, some cationic glycopolymers constructed by amino acids and carbohydrates could act as highly efficient synthetic mimics of natural virus-based gene vectors.36,37 However, up to now, the design and synthesis of well-defined cationic amino acid-based glycopolymer gene carriers with low cytotoxicity, high gene transfection efficiency and preferred serum-compatibility is a challenge. Besides, the structure–activity relationships and the intracellular features (especially precise mechanisms of intracellular uptake and trafficking) of the amino acid-based glycopolymer gene carrier systems are still far from clear and need to be deeply investigated.
To develop new amino acid-based cationic glycopolymers with well-defined structures and elucidate the structure-dependent gene transfection properties, in this work, a series of diblock (PHML-b-PMAGal) and statistical (P(HML-st-MAGal)) cationic glycopolymers with pendant galactose and (L)-lysine moieties were synthesized by RAFT polymerization. The cytotoxicity, in vitro luciferase (Luc) gene transfection efficacy and the impact of serum on the gene transfection efficiency were evaluated in various cell lines. Finally, the cellular uptake/endocytosis pathway and the intracellular localization of the glycopolymers/pDNA payloads were investigated and discussed.
2. Experimental procedures
2.1. Materials
4-Cyano-4-(dodecylsulfanylthiocarbonyl)sulfanyl pentanoic acid was synthesized according to the literature.38 The HEMA-Boc-Lys monomer was synthesized according to our previous work.39 Synthesis of the glyco monomer 6-O-methacryloyl-1,2,3,4-di-O-isopropylidene-galactopyranose (MAIGal) is depicted in S1 (ESI†). Tetrahydrofuran (THF) was dehydrated by refluxing with sodium and purified by distillation before use. 2,2-Azobis(isobutyronitrile) (AIBN) was obtained from Shanghai Sinopharm Chemical Reagent Co., Ltd. (China) and was purified by recrystallization in ethanol. Branched polyethylenimine (bPEI-25k, Mw = 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, Cat#408727) and thiazolyl blue tetrazolium bromide (Cat#M2128) were purchased from Sigma-Aldrich. LysoTracker (Cat#L7526) was purchased from Invitrogen (USA). The Label IT tracker™ intracellular nucleic acid localization kit was purchased from Mirus BioCorporation (Cat#MIR7010, USA). Luciferase assay and bicinchoninic acid (BCA) protein quantitation kits were supplied by Promega (Cat#E1500, USA) and Biomiga (Cat#PW0104, USA), respectively. 0.1 M phosphate buffer solution (PBS 1×), MEM, RPMI-1640 medium and Dulbecco's modified Eagles medium (DMEM) were purchased from Hangzhou Genom Co. Ltd. Fetal bovine serum (FBS) was supplied by GIBCO (Cat#10099-141, USA) and ExCell Bio (Cat#FSP500, China). DAPI was received from Roche (Cat#70217321, Switzerland). The pCMV-Luc plasmid and COS-7 cell lines were generously gifted by Prof. Yuhong Xu of School of Pharmacy of Shanghai Jiaotong University (Shanghai, China). H1299, SK-HEP-1, CHO, HeLa and MCF-7 cell lines were generously gifted by Dr Bo Wan of Fudan University (Shanghai, China). All other chemicals and solvents were of analytical grade and were used as received.
000, Cat#408727) and thiazolyl blue tetrazolium bromide (Cat#M2128) were purchased from Sigma-Aldrich. LysoTracker (Cat#L7526) was purchased from Invitrogen (USA). The Label IT tracker™ intracellular nucleic acid localization kit was purchased from Mirus BioCorporation (Cat#MIR7010, USA). Luciferase assay and bicinchoninic acid (BCA) protein quantitation kits were supplied by Promega (Cat#E1500, USA) and Biomiga (Cat#PW0104, USA), respectively. 0.1 M phosphate buffer solution (PBS 1×), MEM, RPMI-1640 medium and Dulbecco's modified Eagles medium (DMEM) were purchased from Hangzhou Genom Co. Ltd. Fetal bovine serum (FBS) was supplied by GIBCO (Cat#10099-141, USA) and ExCell Bio (Cat#FSP500, China). DAPI was received from Roche (Cat#70217321, Switzerland). The pCMV-Luc plasmid and COS-7 cell lines were generously gifted by Prof. Yuhong Xu of School of Pharmacy of Shanghai Jiaotong University (Shanghai, China). H1299, SK-HEP-1, CHO, HeLa and MCF-7 cell lines were generously gifted by Dr Bo Wan of Fudan University (Shanghai, China). All other chemicals and solvents were of analytical grade and were used as received.
2.2. Preparation of the glycopolymers
1H NMR (300 MHz, CDCl3, δ in ppm): 5.64, 5.15 (NHCOO, 2H, br), 4.48–4.06 (OCH2CH2O, COCH(R) NH, 5H, brm), 3.10 (NHCH2COOR, 2H, m), 2.06–1.49 (CH2, CH2-CCH3, 8H, brm), 1.44 (C(CH3)3, 9H, s), 1.24–0.76 (C-CCH3, 3H, brm).
FTIR (in cm−1): 3380, 2977, 2933, 2867, 1714, 1520, 1456, 1392, 1366, 1250, 1170, 1050, 866, 737, 668.
1H NMR (300 MHz, CDCl3, δ in ppm): 5.68, 5.17 (br, NHCOO), 5.51(s, galactopyranose (Gal)-H at 1 position), 4.63 (m, Gal-H at 3 position), 4.62–3.87 (brm, OCH2CH2O, COCH(R)NH, Gal-H at 2, 4, 5 position), 3.10 (m, NHCH2COOR), 2.06–1.49 (brm, CH2, CH2-CCH3), 1.44 (s, C(CH3)3), 1.24–0.76 (brm, C-CCH3).
FTIR (in cm−1): 3378, 2976, 2932, 1715, 1521, 1456, 1392, 1366, 1250, 1169, 1052, 864, 781.
1H NMR (300 MHz, CDCl3, δ in ppm): 5.68, 5.17 (br, NHCOO), 5.51 [s, galactopyranose (Gal)-H at 1 position], 4.63 (m, Gal-H at 3 position), 4.62–3.87 (brm, OCH2CH2O, COCH(R)NH, Gal-H at 2, 4, 5 position), 3.10 (m, NHCH2COOR), 2.06–1.49 (brm, CH2, CH2-CCH3), 1.44 (s, C(CH3)3), 1.24–0.76 (brm, C-CCH3).
FTIR (in cm−1): 3378, 2976, 2932, 1715, 1521, 1456, 1392, 1366, 1250, 1169, 1052, 864, 781.
PHML: 1H NMR (D2O, δ in ppm): 4.48–3.90 (COOCH2CH2O, COCH(R)NH, 5H, brm), 2.83 (CH2N, 2H, t), 2.02–1.20 (CH2-CCH3, CH2, 8H, brm), 0.94–0.58 (C-CCH3, 3H, d).
PHML-b-PMAGal: 1H NMR (300 MHz, DMSO-d6, δ in ppm): 8.72, 8.08 (s, NH3+CF3COO−), 4.62–3.83 (brm, OCH2CH2O), 3.17 (s, galactose hydroxyl groups), 2.78 (s, –NHCH2COOR), 2.07–1.21 (brm, CH2, CH2-CCH3), 1.04–0.52 (brm, C-CCH3).
FTIR (in cm−1): 3431, 1679, 1539, 1433, 1203, 1136, 966, 839, 801, 723.
P(HML-st-MAGal): 1H NMR (300 MHz, DMSO-d6, δ in ppm): 8.72, 8.08 (s, NH3+CF3COO−), 4.62–3.83 (brm, OCH2CH2O), 3.17 (s, galactose hydroxyl groups), 2.78 (s, –NHCH2COOR), 2.07–1.21 (brm, CH2, CH2-CCH3), 1.04–0.52 (brm, C-CCH3).
FTIR (in cm−1): 3431, 1679, 1539, 1433, 1203, 1136, 966, 839, 801, 723.
2.3. Instrumental characterization of the synthesized cationic polymers
| Polymer samples | M n,NMR /kDa | M n,GPC /kDa | M w/Mn (PDI) | DP (HML) | DP (Gal) | MAGal contc/% |
|---|---|---|---|---|---|---|
| a Molecular weights were estimated by 1H NMR. b Data were obtained by GPC with THF as the eluent, and molecular weights and their distribution were calculated using polystyrene standards. c Weight fraction of the MAGal moiety in the samples was calculated according to the MAIGal cont (%) = [DP(MAIGal) × 248]/Mn,NMR, where 248 expresses the molar mass of the MAGal moiety. | ||||||
| Homopolymer | ||||||
| PHMLBoc40 | 18.6 | 21.1 | 1.19 | 40 | 0 | 0 |
| Block copolymers | ||||||
| PHMLBoc40-b-PMAIGal3 | 19.6 | 28.6 | 1.30 | 40 | 3 | 3.7 |
| PHMLBoc40-b-PMAIGal15 | 23.4 | 34.8 | 1.32 | 40 | 15 | 16.2 |
| Statistical copolymers | ||||||
| P(HMLBoc40-st-MAIGal4) | 19.9 | 29.9 | 1.21 | 40 | 4 | 4.8 |
| P(HMLBoc40-st-MAIGal13) | 22.8 | 31.8 | 1.28 | 40 | 12 | 14.3 |
2.4. Agarose gel electrophoresis
pDNA binding affinity of the PHML, PHML-b-PMAGal and P(HML-st-MAGal) cationic polymers were determined by agarose gel electrophoresis. Each cationic polymer/pDNA polyplex was prepared by mixing cationic polymers with 1 μg pDNA at various N/P charge ratios in deionized water. After 30 min of incubation at 37 °C, the mixed solution was loaded onto agarose gel (1.2% w/v) containing ethidium bromide (0.1 mg mL−1 in gel) in TAE buffer solution, commercially available DNA markers and naked pDNA were used as controls. The electrophoresis was run at 100 V for 40 min. Finally, the pDNA retardation was observed and recorded using a UV transilluminator (UVP, USA).2.5. Average particle size and zeta potential measurements by DLS
Each cationic polymer/pDNA polyplex was prepared by mixing cationic polymers with 1 μg of pDNA (1 mL solution) at various N/P charge ratios in deionized water. Average particle sizes and zeta potential values of the PHML/pDNA, PHML-b-PMAGal/pDNA and P(HML-st-MAGal)/pDNA polyplexes were measured using a dynamic scattering instrument (Malvern Zetasizer Nano ZS90, UK, λ = 633 nm, scattering angle of 90°) at 25 °C.2.6. Transmission electron microscopy (TEM)
The morphology of the PHML40/pDNA, PHML40-b-MAGal3/pDNA and P(HML40-st-MAGal4)/pDNA polyplexes was determined by TEM (JEOL-1230, JEOL Co. Ltd, Japan) at an acceleration voltage of 80 KV. For each sample preparation, the polyplex solution (1.0 mg mL−1) was dropped onto a 300-mesh carbon-coated copper grid, the excess solution was absorbed and removed using filter paper, and the grid was then air-dried at room temperature.2.7. MTT cytotoxicity assay
In vitro cytotoxicity of the PHML, PHML-b-PMAGal and P(HML-st-MAGal) cationic polymers was evaluated by MTT assay in H1299 cells. First, H1299 cells were seeded into 96-well microplates at a density of 5.0 × 103 cells per well in 100 μL of RPMI-1640 medium containing 10% FBS. After 24 h of incubation, the culture medium was replaced with fresh RPMI-1640 medium (100 μL, with 10% FBS), and predetermined amounts of cationic polymers were added and further incubated for another 24 h. Then 20 μL of MTT solution (5 mg mL−1) was added into each well and incubated for additional 2 h at 37 °C. After that, the medium was discarded and 100 μL per well DMSO was added and mildly shaken for 10 min to dissolve the formed MTT formazan. The absorbance of all samples was measured with six replicates (n = 6) by using a microplate reader (BioTek, ELx800, USA) at λ = 490 nm (λ = 630 nm as the reference wavelength). Relative cell viability (%) was calculated according to the equation: relative cell viability (%) = (OD490sample − OD630sample)/(OD490control − OD630control) × 100%.2.8. In vitro luciferase gene transfection assay
For in vitro luciferase gene transfection assay of the PHML, PHML-b-PMAGal and P(HML-st-MAGal) cationic polymers, first, H1299 cells were seeded in 24-well plates (4 × 105 cells per well) with 500 μL of RPMI-1640 medium containing 10% FBS. After incubation for 24 h, the medium were replaced with 500 μL of fresh RPMI-1640 medium (10% FBS) containing the polyplex (1 μg pDNA per well) at various charge ratios and further incubated for 24 h at 37 °C, the commercially available bPEI-25k (N/P = 10) was used as the control. The luciferase assay was performed according to the protocols and relative light units (RLUs) were measured using a Promega GloMax®-Multi Single Tube Multimode Reader (E6070, USA), protein quantification was determined using a BCA assay kit (Applygen Technologies Inc, China) according to the protocol, and relative light units per milligram of luciferase protein (RLU per mg) were calculated to evaluate the cell transfection efficacy of the cationic polymers (n = 3).For in vitro luciferase gene transfection assay of the optimized P(HML40-st-MAGal4)/pDNA polyplexes (N/P = 40) in the presence of serum, the SK-HEP-1, MCF-7, CHO, HeLa and COS-7 cells were separately seeded into 24-well plates (1 × 105 cells per well) with 500 μL of MEM (SK-HEP-1 cells), DMEM (MCF-7, CHO and HeLa cells) or RPMI-1640 (COS-7 cells) medium (all the media contained 10% FBS). After incubation for 24 h, the medium was replaced with fresh medium (+10% FBS) with the P(HML40-st-MAGal4)/pDNA polyplex (1 μg pDNA per well, N/P = 40) and further incubated for 24 h at 37 °C. The luciferase gene expression assay was performed as per the protocols shown above using commercially available bPEI-25k (N/P = 10) as the control.
2.9. Endocytosis mechanism/pathway of the P(HML40-st-MAGal4)/pDNA polyplexes
The endocytosis mechanism/pathway of the P(HML40-st-MAGal4)/pDNA polyplexes was measured by flow cytometry using various endocytosis-specific inhibitors. First, H1299 cells were seeded into 24-well microplates (4 × 105 cells per well) and further cultivated overnight in RPMI 1640 medium containing 10% FBS. Then, the medium in microplates was replaced with fresh RPMI 1640 medium (+10% FBS). Endocytosis inhibitors including amiloride (2.25 μg mL−1, macropinocytosis inhibitor), chlorpromazine (3.0 μg mL−1, clathrin-mediated endocytosis inhibitor), nocodazole (2.0 μg mL−1, microtubule inhibitor) and methyl-β-cyclodextrin (M-β-CD, 2.5 mM, caveolae-mediated endocytosis inhibitor) were, respectively, added into each well and incubated for 1 h. Subsequently, the cells were replaced with fresh RPMI 1640 medium (with 10% FBS) and dealt with the as-prepared P(HML40-st-MAGal4)/pDNA polyplexes (1 μg pDNA per well, N/P = 40). After incubation for 24 h, the luciferase gene expression assay was performed as per the protocols shown above.2.10. Intracellular trafficking of the P(HML40-st-MAGal4)/Cy3-pDNA polyplexes
H1299 cells were seeded into 6-well microplates (4 × 105 cells per well in 1 mL of RPMI 1640 medium with 10% FBS) and incubated at 37 °C and under 5% CO2 for 24 h. The P(HML40-st-MAGal4)/Cy3-pDNA polyplexes were prepared by mixing Cy3-labeled pDNA with the P(HML40-st-MAGal4) polymer (N/P = 40) and incubated at 37 °C for 30 min.35 The as-prepared P(HML40-st-MAGal4)/Cy3-pDNA polyplexes were added into the well and incubated for predetermined time periods (2 h and 6 h). Then the H1299 cells were washed with 1× PBS three times to eliminate fluorescence of the background. The cells were fixed with 4% paraformaldehyde in 0.12 M phosphate buffer (pH = 7.4) for 10 min and washed with 1× PBS three times. LysoTracker (final working concentration 100 ng mL−1) was added to stain the endosome/lysosome and incubated for 15 min before fluorescence imaging. Finally, the intracellular localization of the P(HML40-st-MAGal4)/Cy3-pDNA polyplexes was visualized and recorded on a Nikon Ti–S inverted fluorescence microscope.3. Results and discussion
3.1. Preparation and characterization of the cationic polymers
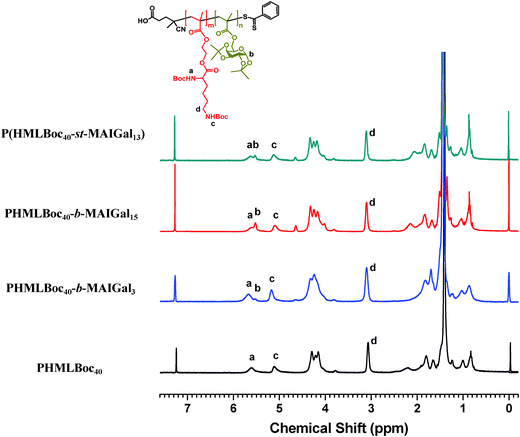
In order to prepare new cationic glycopolymers with the merit of well-defined, functionalized and controllable structures and further study the effect of the functional building blocks and glycopolymer structures (block vs. statistical) on the related physico-chemical and biological properties, a homopolymer PHML with pendent (L)-lysine, and a series of block glycopolymers PHML-b-PMAGal and statistical glycopolymers P(HML-st-MAGal) with pendant galactose and (L)-lysine groups/moieties were prepared via RAFT polymerization (Scheme 1). First, the HEMA-Boc-Lys monomer was synthesized according to our previous work,39 the glyco monomer 6-O-methacryloyl-1,2,3,4-di-O-isopropylidene-galactopyranose (MAIGal) was prepared according to the literature.40 The synthesis routes and characterization of the monomers are shown in the ESI† (Scheme S1 and Fig. S1). The homopolymer PHMLBoc40 was obtained as a precursor via RAFT polymerization of the HEMA-Boc-Lys monomer by using the AIBN/4-cyanopentanoic acid dithiobenzoate system. Then the MAIGal monomer was further polymerized by employing the PHMLBoc40 precursor as a macro-chain-transfer-agent (MacroCTA) and two diblock glycopolymers containing different galactose ratios (PHMLBoc40-b-PMAIGal3 and PHMLBoc40-b-PMAIGal15) were obtained by adjusting the feeding ratio of MAIGal monomers and PHMLBoc40 MacroCTA. For comparison, two statistical glycopolymers P(HMLBoc40-st-MAIGal4) and P(HMLBoc40-st-MAIGal13) were prepared through RAFT polymerization of MAIGal and HEMA-Boc-Lys monomers with predetermined feeding ratios. The structures of these polymers were characterized by 1H NMR (Fig. 1) and GPC (Fig. 2). As shown in Fig. 1, the average degree of polymerization of the MAlGal monomers DP(MAIGal) was calculated via the formula: DP(MAIGal) = (Ia+b − Ic) × 80/Id. As shown in Fig. 2, symmetric peaks were observed with comparatively low molecular weight distribution (Mw/Mn < 1.35), indicating that the synthesized glycopolymers possessed definite molecular structures, which might be beneficial for the elucidation of their chemical structure–biological property relationships. | ||
| Scheme 1 Synthesis of the cationic homopolymer PHML, cationic block glycopolymers PHML-b-PMAGal and cationic statistical glycopolymers P(HML-st-MAGal) via RAFT polymerization and deprotection. | ||
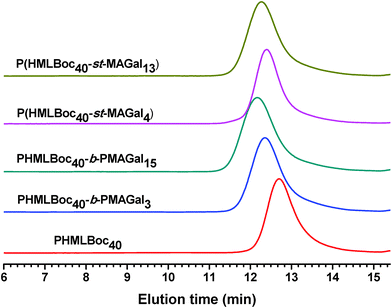 | ||
| Fig. 2 GPC elution traces for the PHMLBoc homopolymer, PHMLBoc-b-PMAIGal block copolymers and P(HMLBoc-st-MAIGal) statistical copolymers. | ||
Finally, the cationic homopolymer PHML, block polymers PHML40-b-PMAGal3 and PHML40-b-PMAGal15, and statistical polymers P(HML40-st-MAGal4) and P(HML40-st-MAGal13) were obtained via TFA-mediated deprotection. The structures of the PHML40-b-MAGal and P(HML40-st-MAGal) cationic polymers are shown in Scheme 1 (the dithiobenzoate in the cationic polymers may be decomposed after TFA-deprotection,41 so the dithiobenzoate end groups were removed and replaced with an asterisk *). Taking P(HML40-st-MAGal13) as an example, the 1H NMR and FTIR characterization are shown in Fig. S2 and S3 (ESI†), respectively. It could be seen that the strong peaks at δ = 1.36–1.42 ppm (acetal and Boc protecting groups) remarkably decreased (Fig. S2, ESI†), and the amide proton signal at 6.6 and 7.0 ppm disappeared along with the appearance of a new protonated amine signal at 8.1 and 8.8 ppm, which indicated the complete removal of the acetal and Boc protecting groups. In Fig. S3 (ESI†), the stretching vibration signal of the methyl group of the acetal and Boc protecting groups (νC–CH3) at 1366 cm−1 disappeared along with the appearance of the hydroxyl group signal (νO–H) at 3431 cm−1, further confirming the structure of P(HML40-st-MAGal13). The results demonstrated the successful preparation of the cationic glycopolymers.
3.2. pDNA binding affinity of the glycopolymers by agarose gel electrophoresis
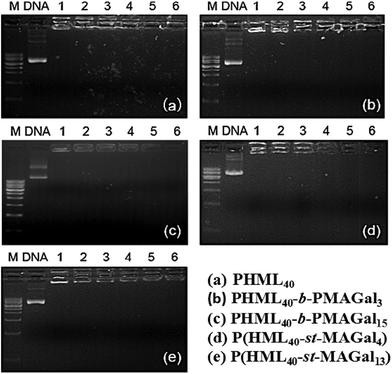
pDNA binding affinities of the synthesized cationic diblock glycopolymers (PHML-b-PMAGal) and statistical glycopolymers (P(HML-st-MAGal)) were evaluated by agarose gel electrophoresis using the cationic homopolymer PHML as the control. As shown in Fig. 3, all the cationic polymers were able to retard pDNA completely at the N/P charge ratio of 1, which might be due to the strong pDNA binding affinity of the primary amino groups (pKa = 10.8) on the (L)-lysine moiety.42 It is noteworthy that different from some PEG-containing cationic polymers, the introduction of galactose moieties into poly-(L)-lysine cationic polymers did not reduce the pDNA binding affinity, which might be partially attributed to the hydrogen bond-mediated interactions between the galactose moiety and pDNA.43 Moreover, the results indicated that the PHML-b-PMAGal and P(HML-st-MAGal) cationic glycopolymers possessed high pDNA binding affinity, which made them efficient cargoes for pDNA loading.3.3. Average particle size and zeta potential measurements
The average particle size and zeta potential of polymers/pDNA polyplexes were essential factors for evaluating the cytotoxicity, cellular uptake and gene transfection efficiency.34 Herein, the average particle sizes of the cationic glycopolymers/pDNA polyplexes at various N/P charge ratios were measured by DLS using the homopolymer PHML as control. As shown in Fig. 4, all the polyplexes displayed an average size range of 100–300 nm within the N/P charge ratio of 10–100. It can be found that the PHML40-b-PMAGal15/pDNA and P(HML40-st-MAGal13)/pDNA (N/P = 40) polyplexes, which contain a higher percentage of galactose moieties (16.2% and 14.3%, respectively), showed a comparatively larger particle size than the other polyplexes, which might be due to the formation of a galactose-containing hydrophilic shell/corona on the surface of polyplex payloads by the strong hydration effect of galactose moieties. Moreover, morphology of the PHML40/pDNA, PHML40-b-MAGal3/pDNA and P(HML40-st-MAGal4)/pDNA polyplexes was examined by transmission electron microscopy (TEM). As shown in Fig. S4 (ESI†), all of the polyplexes formed spherical-shaped nanoparticles with the size range of 30–110 nm, which was smaller than the size measured by DLS, due to the shrinkage of the nanoparticles in a drying process during the preparation of TEM samples.35,44 Besides, the zeta potentials of the polyplexes were measured by DLS. As shown in Fig. 4b, the polyplexes displayed positive surface potentials (+30.2 to +46.3 mV) at N/P charge ratios from 10 to 100, for each polyplex, the zeta potential was almost maintained at the same level and showed less dependence of the N/P charge ratios (N/P 10–100), suggesting the complete pDNA binding of the cationic polymers within the N/P range, due to the strong electrostatic interactions between the cationic lysine groups/moieties and pDNA, which is in accordance with the above electrophoresis results and our previous work.39 Moreover, the relatively small particle size and the positive surface potential of the polyplexes may be beneficial to their cell surface adhesion and the following intracellular uptake process,36 which may further improve the intracellular gene transfection capability.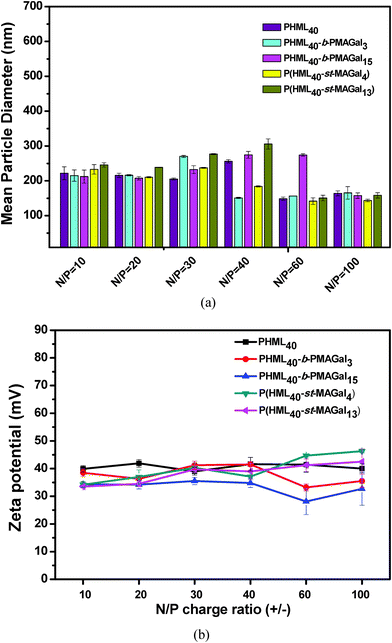 | ||
| Fig. 4 (a) Particle size and (b) zeta potential of the PHML, PHML-b-PMAGal and P(HML-st-MAGal)/pDNA polyplexes at various N/P ratios determined by DLS. | ||
3.4. Cytotoxicity
Cytotoxicity is one of the most important parameters for evaluating the safety of the non-viral gene vectors toward therapeutic applications. Recently, it has been revealed that introducing some natural moieties such as cholesterol, sugar and amino acids could improve the biocompatibility and decrease the cytotoxicity of polymeric gene vectors.34 Herein, the cytotoxicity of cationic polymers PHML, PHML-b-PMAGal and P(HML-st-MAGal) was measured by MTT assay using the commercially available bPEI-25k as control. As shown in Fig. 5, severe cytotoxicity (relative cell viability, RCV < 30%) was observed in H1299 cells when incubated with bPEI-25k at the dose of up to 60 μg mL−1, whereas PHML, PHML40-b-PMAGal3 and P(HML40-st-MAGal4) showed lower cytotoxicity (RCV > 60%) than bPEI-25k in the same dose range. It is noteworthy that the galactose-rich cationic polymers PHML40-b-PMAGal15 and P(HML40-st-MAGal13) exhibited much lower cytotoxicity (RCV > 80%) even at a high concentration of 200 μg mL−1, which indicated that introducing biocompatible galactose moieties could decrease the cytotoxicity of cationic polymers. Similarly, previous literature studies had reported that sugar (cyclodextrin)-conjugation on some cationic dendrimer gene carriers could lower their cytotoxicity.45 Moreover, the MTT results suggested that the cationic glycopolymers might be employed as comparatively safe vectors for gene delivery applications.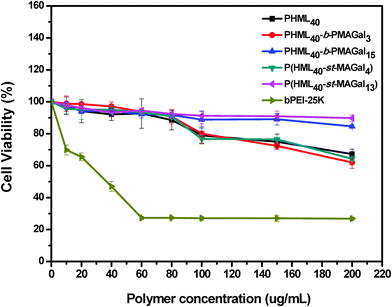 | ||
| Fig. 5 MTT cytotoxicity assay of the cationic polymers PHML, PHML-b-PMAGal and P(HML-st-MAGal) in H1299 cells at various dosages. | ||
3.5. In vitro luciferase gene transfection
The in vitro gene transfection activity of PHML, PHML-b-PMAGals and P(HML-st-MAGal)s cationic polymers was evaluated in the presence of serum (10% FBS) in H1299 cells, using commercially available “gold standard” bPEI-25k as the control. As shown in Fig. 6, the optimum transfection efficiency of the homopolymer PHML is 4.5-fold higher than that of bPEI-25k, and the low-galactose-content cationic glycopolymers PHML40-b-PMAGal3 and P(HML40-st-MAGal4) showed their highest transfection efficiency at the N/P ratio of 40, 1.4 to 6.8-fold higher than the transfection level of bPEI-25k (**P < 0.05). However, the high-galactose-content cationic glycopolymers PHML40-b-PMAGal15 and P(HML40-st-MAGal13) showed comparatively lower transfection efficiencies than their low-galactose-content counterparts. The lower gene transfection efficiency of the high-galactose-content cationic polymers might be due to the surface charge shielding effect of their excess galactose moieties, which hindered the effective adhesion of the cationic lysine moieties to the negatively-charged cell membrane, thus further decreasing the cellular uptake capability and subsequent gene expression. It is noteworthy that the statistical glycopolymer P(HML40-st-MAGal4) showed a higher transfection capability than the block glycopolymer PHML40-b-PMAGal3, which has a similar chemical component but a different copolymer sequence/arrangement, indicating that the gene transfection efficiency depends to some extent on the copolymer sequence/arrangement of the cationic glycopolymers. Similarly, Ahmed et al.29 observed a similar transfection trend on a series of glucose-containing cationic glycopolymers; they supposed that the gene transfection difference might be attributed to the different interactions between the sequence-varied cationic glycopolymers and the negatively-charged serum proteins.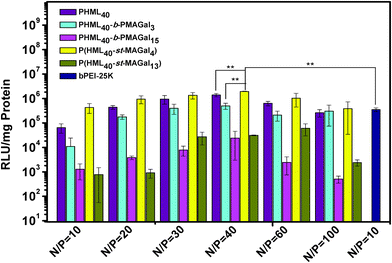 | ||
| Fig. 6 Luciferase gene transfection efficiency of the PHML, PHML-b-PMAGal and P(HML-st-MAGal) vectors at different N/P ratios with H1299 cells in the presence of 10% FBS (**P < 0.05). | ||
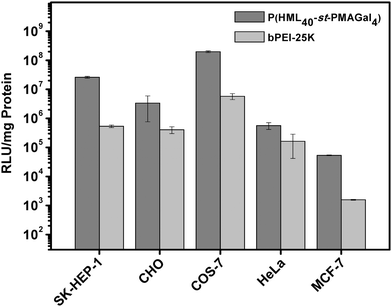
To further evaluate the practical applications of the cationic glycopolymers in different cell lines under serum conditions (10% FBS), based on the transfection results in H1299 cells, the optimized statistical cationic glycopolymer P(HML40-st-MAGal4) polyplexes (N/P = 40) were utilized as the transfection model. As shown in Fig. 7, the P(HML40-st-MAGal4) polyplexes showed obviously higher luciferase gene transfection efficiency (4.8–87-fold) than the “gold standard” bPEI-25k in SK-HEP-1, MCF-7, CHO, HeLa and COS-7 cells. The results suggested that the statistical cationic glycopolymer P(HML40-st-MAGal4) might be employed as a serum-compatible and highly efficient transfection agent for cellular gene delivery and practical gene therapy.
3.6. Endocytosis mechanism/pathway of the P(HML40-st-MAGal4)/pDNA polyplexes
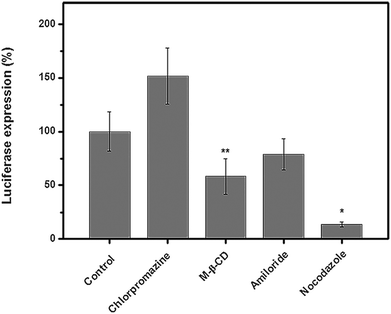
It had been revealed that the gene/drug delivery efficiency of nano-delivery systems greatly relied upon their endocytosis mechanism/pathway;46 to our knowledge, studies on the endocytosis pathway of cationic glycopolymer vectors/pDNA complexes are still scarce. Herein, in order to elucidate the endocytosis pathway of the P(HML40-st-MAGal4)/pDNA polyplexes (N/P = 40, optimized condition) in the serum environment (10% FBS), we investigated the luciferase expression in the presence of various endocytosis-specific inhibitors.47 The H1299 cells transfected with P(HML40-st-MAGal4)/pDNA polyplexes (N/P = 40) in the absence of inhibitors was set as the control (luciferase expression: 100%). As shown in Fig. 8, the luciferase expression of the P(HML40-st-MAGal4)/pDNA polyplexes obviously decreased with the addition of M-β-CD (62.3%) and nocodazole (18.2%), respectively, indicating that the P(HML40-st-MAGal4)/pDNA polyplexes entered H1299 cells mainly through the caveolae-mediated (or lipid-raft-mediated) endocytosis and the microtubule-dependent pathway. Interestingly, with addition of the clathrin-mediated endocytosis inhibitor chlorpromazine, the luciferase tranfection increased to around 150% (1.5-fold). Braeckmans et al.48 also reported similar results, which might be attributed to the proton-buffering effect of chlorpromazine. The related mechanism details are not very clear until now. Moreover, the luciferase expression of the H1299 cells incubated with the P(HML40-st-MAGal4)/pDNA polyplexes decreased to 86.2% with addition of the macropinocytosis inhibitor amiloride, which indicated that macropinocytosis might not serve as the dominant endocytosis pathway. In previous works, we disclosed that a series of arginine-bearing polymers with high gene transfection efficiency undergo the caveolae (lipid-raft) mediated endocytosis pathway followed by endosome localization.47 Similarly, Helenius et al.49 revealed that caveosome–endoplasmic reticulum localization was followed by caveolae-mediated endocytosis of the cationic cargoes. Thus, we deduced that the caveolae-mediated (or lipid-raft-mediated) endocytosis pathway may contribute to the high transfection efficiency of the P(HML40-st-MAGal4)/pDNA polyplexes.3.7. Intracellular trafficking and localization of the P(HML40-st-MAGal4)/pDNA polyplexes
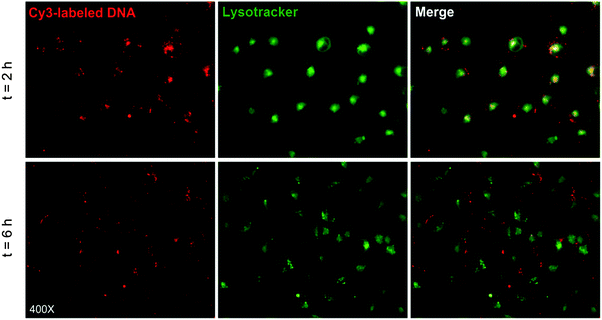
In order to further explore the transfection mechanism, the intracellular trafficking and localization of the P(HML40-st-MAGal4)/Cy3-pDNA polyplexes in the presence of serum (10% FBS) were investigated using fluorescence microscopy in H1299 cells, and the endosomes/lysosomes of H1299 cells were stained by the LysoTracker kit.50 As shown in Fig. 9 (merged image), after transfection for 2 h, it could be observed that the red fluorescence of P(HML40-st-MAGal4)/Cy3-pDNA polyplexes was overlapped by the green fluorescence of LysoTracker and resulted in an obvious yellow fluorescence emission, indicating that the polyplexes were entrapped in endosomes/lysosomes. After 6 h of incubation, the yellow fluorescence almost disappeared and the red and green fluorescence dots clearly separated, which indicated the successful and efficient “lysosome escape” effect. The results might be attributed to the proton buffering effect of cationic (L)-lysine moieties, which was regarded as one of the crucial factors to achieve efficient gene transfection. Moreover, some of the polyplexes could be found distributed inside the H1299 cell nuclei (Fig. S5, ESI†). Recently, it has been reported that internalized sugar-containing nanoparticles demonstrated the “Trojan horse effect” and were able to colocalize within various subcellular organelles such as the endosome/lysosome, endoplasmic reticulum, Golgi apparatus, and so on.51 Based on this, we expected that the cationic glycopolymers with certain sugar-moieties may possess a sugar-mediated intracellular trafficking/localization effect, which may further provide new approaches for improving the gene/drug delivery efficacy by directing gene/drug payloads to go through the probably efficient “sugar-mediated pathways”. In addition, further research on the correlation between sugar/carbohydrate moieties and the intracellular localization/trafficking characteristics of the glycopolymer-based gene/drug delivery systems is currently underway in our lab.4. Conclusions
In summary, a series of block/statistical cationic glycopolymers PHML-b-PMAGal and P(HML-st-MAGal) with pendant (L)-lysine and galactose moieties were prepared via RAFT polymerization followed by Boc-deprotection. All the synthesized cationic glycopolymers showed high pDNA binding affinity at a low N/P charge ratio of 1. The average hydrodynamic particle diameter (150–300 nm) and the positive zeta potential (+30–45 mV) of the cationic glycopolymers/pDNA polyplexes were found suitable for the intracellular uptake. MTT assay showed that the cationic glycopolymers had significantly lower cytotoxicity than bPEI-25k, which depended on the block/statistical polymer architecture and the galactose content. Moreover, it could be noticed that the statistical copolymer P(HML40-st-MAGal4) with 4.8% galactose content showed the highest gene transfection efficiency among the synthesized cationic glycopolymers in the presence of 10% FBS, much higher than the “gold standard” bPEI-25k in various (H1299, SK-HEP-1, MCF-7, CHO, HeLa and COS-7) cell lines. Endocytosis pathway analysis indicated that the P(HML40-st-MAGal4)/pDNA polyplexes entered H1299 cells mainly through the caveolae-mediated (or lipid-raft-mediated) endocytosis pathway and greatly depended on microtubules. Moreover, the P(HML40-st-MAGal4)/pDNA polyplexes showed relatively fast cellular uptake capability and an obvious endosome/lysosome escaping effect. In conclusion, the current results suggested that the synthesized cationic glycopolymers might serve as potential candidates for safe and efficient gene delivery in practical applications.Acknowledgements
The authors are indebted to the partial financial support from the National Science Foundation of China (21174160, 21002116 and 21372251). The authors are also grateful to Prof. Xiangyang Shi and Dr Xueyan Cao of Donghua University for their kind assistance in FACS measurements. Dr Ruilong Sheng appreciates the Chinese Academy of Sciences (CAS) for the CAS-Canada Young Visiting Scientist Scholarship and Youth Innovation Promotion Association (YIPA) for the sponsorship. We also thank the senior scientist Dr Wilms Baille from Université de Montréal for his help on the presentations in the manuscript.References
- D. He and E. Wagner, Macromol. Biosci., 2015, 15, 600–612 CrossRef CAS PubMed.
- D. W. Pack, A. S. Hoffman, S. Pun and P. S. Stayton, Nat. Rev. Drug Discovery, 2005, 4, 581–593 CrossRef CAS PubMed.
- M. A. Mintzer and E. E. Simanek, Chem. Rev., 2009, 109, 259–302 CrossRef CAS PubMed.
- G. Lin, H. Zhang and L. Huang, Mol. Pharmaceutics, 2015, 12, 314–321 CrossRef CAS PubMed.
- J. Dai, S. Zou, Y. Pei, D. Cheng, H. Ai and X. Shuai, Biomaterials, 2011, 32, 1694–1705 CrossRef CAS PubMed.
- S. K. Tripathy, H. B. Black, E. Goldwasser and J. M. Leiden, Nat. Med., 1996, 2, 545–550 CrossRef CAS PubMed.
- S. Son and W. J. Kim, Biomaterials, 2010, 31, 133–143 CrossRef CAS PubMed.
- A. Abe, A. Miyanohara and T. Friedmann, Gene Ther., 1998, 5, 708–711 CrossRef CAS PubMed.
- R. Tang, R. N. Palumbo, L. Nagarajan, E. Krogstad and C. Wang, J. Controlled Release, 2010, 142, 229–237 CrossRef CAS PubMed.
- H. Petersen, T. Merdan, K. Kunath, D. Fischer and T. Kissel, Bioconjugate Chem., 2002, 13, 812–821 CrossRef CAS PubMed.
- K. Knop, R. Hoogenboom, D. Fischer and U. S. Schubert, Angew. Chem., Int. Ed., 2010, 49, 6288–6308 CrossRef CAS PubMed.
- F. Dai and W. Liu, Biomaterials, 2011, 32, 628–638 CrossRef CAS PubMed.
- Y. T. Chim, J. K. Lam, Y. Ma, S. P. Armes, A. L. Lewis, C. J. Roberts, S. Stolnik, S. J. Tendler and M. C. Davies, Langmuir, 2005, 21, 3591–3598 CrossRef CAS PubMed.
- A. E. Smith, A. Sizovs, G. Grandinetti, L. Xue and T. M. Reineke, Biomacromolecules, 2011, 12, 3015–3022 CrossRef CAS PubMed.
- S. P. Strand, M. M. Issa, B. E. Christensen, K. M. Varum and P. Artursson, Biomacromolecules, 2008, 9, 3268–3276 CrossRef CAS PubMed.
- Y. Liu and T. M. Reineke, Biomacromolecules, 2010, 11, 316–325 CrossRef CAS PubMed.
- Y. Liu, L. Wenning, M. Lynch and T. M. Reineke, J. Am. Chem. Soc., 2004, 126, 7422–7423 CrossRef CAS PubMed.
- Y. Liu and T. M. Reineke, Bioconjugate Chem., 2007, 18, 19–30 CrossRef CAS PubMed.
- S. R. S. Ting, G. Chen and M. H. Stenzel, Polym. Chem., 2010, 1, 1392–1412 RSC.
- B. Voit and D. Appelhans, Macromol. Chem. Phys., 2010, 211, 727–735 CrossRef CAS.
- T. Azzam, A. Raskin, A. Makovitzki, H. Brem, P. Vierling, M. Lineal and A. J. Domb, Macromolecules, 2002, 35, 9947–9953 CrossRef CAS.
- S. R. Popielarski, S. Mishra and M. E. Davis, Bioconjugate Chem., 2003, 14, 672–678 CrossRef CAS PubMed.
- J. Chang, X. Xu, H. Li, Y. Jian, G. Wang, B. He and Z. Gu, Adv. Funct. Mater., 2013, 23, 2691–2699 CrossRef CAS.
- K. Nomura and R. R. Schrock, Macromolecules, 1996, 29, 540–545 CrossRef CAS.
- J. Bernard, X. Hao, T. P. Davis, C. Barner-Kowollik and M. H. Stenzel, Biomacromolecules, 2006, 7, 232–238 CrossRef CAS PubMed.
- M. Ahmed and R. Narain, Biomaterials, 2013, 34, 4368–4376 CrossRef CAS PubMed.
- M. Ahmed and R. Narain, Biomaterials, 2012, 33, 3990–4001 CrossRef CAS PubMed.
- M. Ahmed, M. Jawanda, K. Ishihara and R. Narain, Biomaterials, 2012, 33, 7858–7870 CrossRef CAS PubMed.
- M. Ahmed and R. Narain, Biomaterials, 2011, 32, 5279–5290 CrossRef CAS PubMed.
- D. Sprouse and T. M. Reineke, Biomacromolecules, 2014, 15, 2616–2628 CrossRef CAS PubMed.
- Y. Wu, M. Wang, D. Sprouse, A. E. Smith and T. M. Reineke, Biomacromolecules, 2014, 15, 1716–1726 CrossRef CAS PubMed.
- L. Xue, N. P. Ingle and T. M. Reineke, Biomacromolecules, 2013, 14, 3903–3915 CrossRef CAS PubMed.
- V. P. Taori, H. Lu and T. M. Reineke, Biomacromolecules, 2011, 12, 2055–2063 CrossRef CAS PubMed.
- R. Sheng, T. Luo, H. Li, J. Sun, Z. Wang and A. Cao, Colloids Surf., B, 2014, 116, 32–40 CrossRef CAS PubMed.
- R. Sheng, T. Luo, Y. Zhu, H. Li, J. Sun, S. Chen, W. Sun and A. Cao, Biomaterials, 2011, 32, 3507–3519 CrossRef CAS PubMed.
- X. Xu, H. Yuan, J. Chang, B. He and Z. Gu, Angew. Chem., Int. Ed., 2012, 51, 3130–3133 CrossRef CAS PubMed.
- W.-D. Jang, K. M. Kamruzzaman Selim, C.-H. Lee and I.-K. Kang, Prog. Polym. Sci., 2009, 34, 1–23 CrossRef CAS.
- G. Moad, Y. K. Chong, A. Postma, E. Rizzardo and S. H. Thang, Polymer, 2005, 46, 8458–8468 CrossRef CAS.
- J. Sun, T. Luo, R. Sheng, H. Li, S. Chen, F. Hu and A. Cao, Macromol. Biosci., 2013, 13, 35–47 CrossRef CAS PubMed.
- F. Suriano, O. Coulembier, P. Degée and P. Dubois, J. Polym. Sci., Part A: Polym. Chem., 2008, 46, 3662–3672 CrossRef CAS.
- J. Lu, C. Fu, S. Wang, L. Tao, L. Yan, D. Haddleton, G. Chen and Y. Wei, Macromolecules, 2014, 47, 4676–4683 CrossRef CAS.
- A. M. Funhoff, C. F. van Nostrum, G. A. Koning, N. M. E. Schuurmans-Nieuwenbroek, D. J. A. Crommelin and W. E. Hennink, Biomacromolecules, 2004, 5, 32–39 CrossRef CAS PubMed.
- Y. Miura, Polym. J., 2012, 44, 679–689 CrossRef CAS.
- R. Sheng, F. An, Z. Wang, M. Li and A. Cao, RSC Adv., 2015, 5, 12338–12345 RSC.
- H. Arima, K. Motoyama and T. Higashi, Adv. Drug Delivery Rev., 2013, 65, 1204–1214 CrossRef CAS PubMed.
- S. Zhang, H. Gao and G. Bao, ACS Nano, 2015, 9, 8655–8671 CrossRef CAS PubMed.
- H. Li, T. Luo, R. Sheng, J. Sun, Z. Wang and A. Cao, Biomaterials, 2013, 34, 7923–7938 CrossRef CAS PubMed.
- D. Vercauteren, R. E. Vandenbroucke, A. T. Jones, J. Rejman, J. Demeester, S. C. De Smedt, N. N. Sanders and K. Braeckmans, Mol. Ther., 2010, 18, 561–569 CrossRef CAS PubMed.
- L. Pelkmans, J. Kartenbeck and A. Helenius, Nat. Cell Biol., 2001, 3, 473–483 CrossRef CAS PubMed.
- R. Sheng, T. Luo, H. Li, J. Sun, Z. Wang and A. Cao, Bioorg. Med. Chem., 2013, 21, 6366–6377 CrossRef CAS PubMed.
- D. Benito-Alifonso, S. Tremel, B. Hou, H. Lockyear, J. Mantell, D. J. Fermin, P. Verkade, M. Berry and M. C. Galan, Angew. Chem., Int. Ed., 2014, 53, 810–814 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6tb00969g |
| This journal is © The Royal Society of Chemistry 2016 |