Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Fabrication of living soft matter by symbiotic growth of unicellular microorganisms†
Anupam A. K.
Das
a,
James
Bovill
a,
Maram
Ayesh
a,
Simeon D.
Stoyanov
bc and
Vesselin N.
Paunov
*a
aDepartment of Chemistry, University of Hull, Hull, UK. E-mail: v.n.paunov@hull.ac.uk; Tel: +44 (0)1482 465660
bLaboratory of Physical Chemistry and Soft Matter, Wageningen University, 6703 HB Wageningen, The Netherlands
cDepartment of Mechanical Engineering, University College London, Torrington Place, London WC1E 7JE, UK
First published on 3rd May 2016
Abstract
We report the fabrication of living soft matter made as a result of the symbiotic relationship of two unicellular microorganisms. The material is composed of bacterial cellulose produced in situ by acetobacter (Acetobacter aceti NCIMB 8132) in the presence of photosynthetic microalgae (Chlamydomonas reinhardtii cc-124), which integrates into a symbiotic consortium and gets embedded in the produced cellulose composite. The same concept of growing living materials can be applied to other symbiotic microorganism pairs similar to the combination of algae and fungi in lichens, which is widespread in Nature. We demonstrate the in situ growth and immobilisation of the C. reinhardtii cells in the bacterial cellulose matrix produced by the simultaneous growth of acetobacter. The effect of the growth media composition on the produced living materials was investigated. The microstructure and the morphology of the produced living biomaterials were dependent on the shape of the growth culture container and media stirring conditions, which control the access to oxygen. As the photosynthetic C. reinhardtii cells remain viable and produce oxygen as they spontaneously integrate into the matrix of the bacterial cellulose generated by the acetobacter, such living materials have the potential for various applications in bio-hydrogen generation from the immobilised microalgae. The proposed approach for building living soft matter can provide new ways of immobilising other commercially important microorganisms in a bacterial cellulose matrix as a result of symbiosis with acetobacter without the use of synthetic binding agents and in turn increase their production efficiency.
Introduction
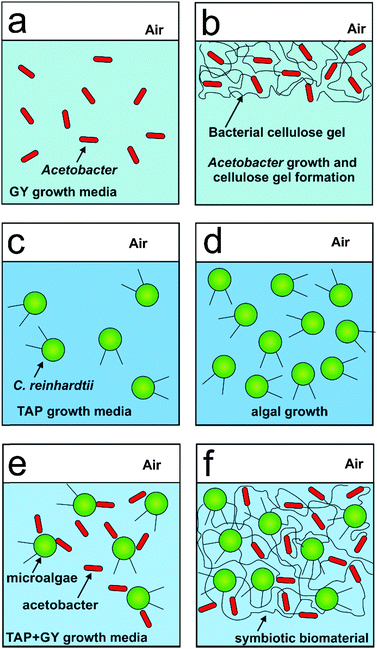
Symbiosis is the relationship between two or more organisms involved to create conditions favourable for mutual growth and proliferation of each other with important implications in the ecology of the system.1 Lichens are one of the most well-studied examples of symbiosis to exist naturally in the environment.2–4 They consist mainly of a filamentous fungus (mycobiont) and a photosynthesising alga or a cyanobacterium (photobiont).5,6 The interactions between the two organisms are not random and depend on various environmental parameters. Lichens can survive in harsher environments compared to the constituting fungi and algae as individual living microorganisms thanks to passing nutrients and metabolites to each other which ensures better adaptability of the lichen.7 In this study, we are aiming to develop a new concept for the preparation of living soft matter by mimicking the relationship between two different microorganisms living symbiotically. Two different classes of microorganisms were grown together in a specific medium where one of the cultures produces the substrate for the growth and proliferation of the other culture, which in turn uses the metabolites produced by the first microorganism and produces other common metabolites. There is evidence that bacterial species can be associated with microalgal species,8 which suggests that there are specific mechanisms of interaction which benefit both the microalgae and the bacteria to enable their coexistence in the same habitat. There are other examples, where Pseudomonas sp., a gram negative bacterium can grow symbiotically along with Chlorella vulgaris.9In this study, we explore the symbiotic combination of the acetobacter (Acetobacter aceti NCIMB 8132) and a photosynthesising microalga (Chlamydomonas reinhardtii cc-124). Our aim is to use their symbiotic relationship to fabricate a new type of living biomaterial (tissue) by growing the microalgae in the same culture media with the acetobacter and immobilise their cells in situ in the bacterial cellulose gel which is one of the by-products of the acetobacter metabolism.
Previous research has shown that many different cultures can be immobilised or grown on already produced bacterial cellulose (nata de coco). Other microorganisms have been successfully immobilised on bacterial cellulose for efficient generation of various kinds of by-products. Yeast has been grown on bacterial cellulose to increase the wine production efficiency and improve the process economy.10 Cellulose fibres produced by Acetobacter xylinum were used to immobilise Pseudomonas Stutzeri for the process of biological denitrification of waste water for the removal of nitrates and their conversion into nitrogen gas.11 The bacterial cellulose made by Glucanobacter xylinus was shown to be highly biocompatible by growing human osteoblast cells with rapid attachment and extension.12 In our case, the immobilisation of the microalgae was done in real time during the production of the bacterial cellulose gel along with a symbiotic relationship with the acetobacter which results in the growth and proliferation of the living biomaterial. It has been previously reported that the microalgae C. reinhardtii have the ability to produce biohydrogen in suspension culture as well as immobilised culture with the latter found to be more efficient,13 hence the living soft matter may have advantages like improved hydrogen production from immobilised culture and various other metabolite extractions.
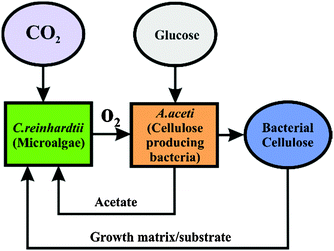
Here we immobilised the microalgae cells in real time with the bacterial cellulose produced by the acetobacter in the same culture consortia. We report the production of a new living soft matter using two different unicellular microorganisms microalgae and cellulose producing bacteria which normally do not form colonies as single cultures. The acetobacter cells are aerobes that use sugars with the production of cellulose and acetic acid as their major byproducts.14 This bacterial cellulose matrix was simultaneously used as a substrate for the growth of the photosynthesising microalgae C. reinhardtii. The microalgae in turn provide oxygen and pass it onto the cellulose producing bacteria (see Scheme 1) in their environment. This allowed the acetobacter to produce cellulose not only at the air–water interface but also around the photosynthetic microalgae which get integrated in the cellulose gel. The schematics of the metabolite exchange during the symbiotic growth is shown in Fig. 1.
The paper is organised as follows. We study the optimal composition of the media which favours symbiotic growth of the living soft matter. The viability of the integrated microalgae and the microstructure of the produced living materials under different growth media conditions is also examined. We monitor the media acidity during the symbiotic growth of the living materials at different initial ratios of the two cultures. We discuss the similarities with lichens in Nature and the possible application of this new approach for growing other types of living materials.
Materials and methods
Materials
Microalgae cultures were grown in the tris-acetate-phosphate (TAP) culture medium at an incubation temperature of 30 °C. The culture medium of C. reinhardtii consisted of TAP salts (ammonium chloride, NH4Cl; magnesium sulphate, MgSO4·7H2O and calcium chloride CaCl2·2H2O), phosphate buffer solution and Hutner's trace element solution (EDTA disodium salt, ZnSO4·7H2O, H3BO3, MnCl2·4H2O, CoCl2·6H2O, CuSO4·5H2O, FeSO4·7H2O, (NH4)6Mo7O24·4H2O, all purchased from Sigma-Aldrich, UK). Acetobacter cultures were grown in glucose/yeast extract (GY) media at an incubation temperature of 28 °C. The acetobacter growth media consist of glucose purchased from Fisher Scientific, UK and yeast extract purchased from Oxoid Ltd, UK. The fluorescein diacetate (FDA) assay used to determine the viability of the C. reinhardtii cell was purchased from Sigma Aldrich, UK. Deionised water produced using a Milli-Q reverse osmosis system (Millipore, UK) was used in all experiments.Algal strain and growth conditions
The C. reinhardtii cc-124 strain was kindly provided by the research group of Prof Michael Flickinger at North Carolina State University, USA. The cells were grown photoheterotrophically in acetate-rich medium in 100, 250 and 500 mL Erlenmeyer flasks containing 50, 150 and 250 mL of standard tris-acetate-phosphate (TAP + S) medium at pH 7.0.15 The flask was placed in a water jacket connected to a thermostatic water bath maintained at 25–30 °C and placed on a magnetic stirrer with a stirring bar to keep the culture agitated at 500 rpm. The setup was illuminated using two 23 W cool white lights (2800 Lux) providing photosynthetically active radiation of 36.4 μE m−2 s−1. The cells were grown up to three days and harvested before their late logarithmic growth phase by centrifugation at 3000 rpm for 5 min. All glassware was autoclaved at 121 °C for 20 min before use for cell growth.Acetobacter strain and growth conditions
The A. aceti (NCIMB 8132) was purchased from NCIMB Ltd, UK. The cells were grown in glucose and yeast extract media (GY media) in 250 mL Erlenmeyer flasks containing 150 mL of GY media.16 The flask was placed in a Stuart orbital incubator SI500 maintained at 28 °C. The cells were grown for up to seven days along with the formation of different layers of cellulose produced by the acetobacter.Microalgae immobilisation procedure and symbiosis
The microalgae (C. reinhardtii) were immobilised in the bacterial cellulose gel produced by A. aceti. The cells were grown together in a mixture of TAP and GY media at various concentrations of 100% GY media (only A. aceti), 100% TAP media (only C. reinhardtii), 100% GY media (with C. reinhardtii and A. aceti), 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 GY
10 GY![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TAP media (v/v), 80
TAP media (v/v), 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 GY
20 GY![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TAP media (v/v), and 70
TAP media (v/v), and 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 GY
30 GY![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TAP media (v/v). The initial inoculum amounts and their ratio were also varied for both microorganisms in the mixture as shown in Tables 1 and 2. The microalgae cells integrated into the bacterial cellulose gel composite were formed in different culture containers, such as a beaker, conical flask, Petri dish and leaf-shaped container, in order to mould the living soft matter into various sizes and shapes. The pH of the culture media was measured at various times during the symbiotic growth using a Fisher brand Hydrus 300 pH meter connected to a Jenway electrode.
TAP media (v/v). The initial inoculum amounts and their ratio were also varied for both microorganisms in the mixture as shown in Tables 1 and 2. The microalgae cells integrated into the bacterial cellulose gel composite were formed in different culture containers, such as a beaker, conical flask, Petri dish and leaf-shaped container, in order to mould the living soft matter into various sizes and shapes. The pH of the culture media was measured at various times during the symbiotic growth using a Fisher brand Hydrus 300 pH meter connected to a Jenway electrode.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 GY
30 GY![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TAP media (v/v) type containing different amounts of C. reinhardtii (grams of wet weight) and A. aceti (mL of broth) used for reference to the graphs shown in Fig. 4
TAP media (v/v) type containing different amounts of C. reinhardtii (grams of wet weight) and A. aceti (mL of broth) used for reference to the graphs shown in Fig. 4
| Sample | A. aceti (mL) | C. reinhardtii (g) |
|---|---|---|
| 1 | 2 | 0.35 |
| 2 | 4 | 0.35 |
| 3 | 6 | 0.35 |
| 4 | 12 | 0.35 |
| 5 | — | 0.5 |
| 6 | 10 | — |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 GY
30 GY![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TAP media (v/v) type containing different amounts of C. reinhardtii (grams of wet weight) and A. aceti (mL of broth) used for reference to the graphs shown in Fig. 5
TAP media (v/v) type containing different amounts of C. reinhardtii (grams of wet weight) and A. aceti (mL of broth) used for reference to the graphs shown in Fig. 5
| Sample | A. aceti (mL) | C. reinhardtii (g) |
|---|---|---|
| 7 | 10 | 0.35 |
| 8 | 10 | 0.45 |
| 9 | 10 | 0.55 |
| 10 | 10 | 0.60 |
| 5 | — | 0.5 |
| 6 | 10 | — |
Microscopy studies of the living soft matter
The viability of C. reinhardtii cells was monitored using a BX-51 fluorescence microscope equipped with a DP70 digital camera and fluorescein diacetate (FDA) as a live/dead cell assay. Fluorescein diacetate (FDA) is a non-fluorescent compound which easily diffuses into intact cells.17,18The result is the uptake and hydrolysis of the non-fluorescent precursor (FDA) by esterase inside the cell and accumulation of a fluorescent byproduct (fluorescein).19 We used 0.5% (w/v) FDA dissolved in acetone. 20 μL of this solution was used per mL of the cell sample. The tube with the mixture was shaken for 10 minutes using a vortex at 1500 rpm under dark conditions to avoid the photobleaching of the produced fluorescein as a result of the enzymatic hydrolysis of FDA on the inside of the viable cells. The sample was washed three times with Milli-Q water using a centrifuge (Eppendorf Minispin plus) at 3500 rpm for 5 minutes in order to remove any extracellular fluorescein. The SEM images were obtained using a Zeiss Evo 60 Scanning Electron Microscope. The living soft matter samples were fixed using 2.5% glutaraldehyde in phosphate buffer followed by incubation in 1% osmium tetroxide in phosphate buffer for 1 hour. They were then gradually dehydrated in ethanol (30%, 50%, 70%, 90% and two times dry 100%) before being dried (at the critical point) using liquid carbon dioxide in a critical point dryer B7010. The samples were coated with Au/Pd after fixing using a Polaron Sputter Coater, model SC7640 fitted with a coating thickness monitor.
They were mounted on aluminium SEM stubs and evacuated inside the coater using a rotary vacuum pump. Vacuum was regulated with a leak valve connected to an argon supply system. The operating pressure was 2–4 Pa. A direct current voltage (about 1 kV) was applied to the source material (Au/Pd target), which ionises the argon gas. Argon ions were accelerated towards the target, ejecting metal atoms in the process which coat the samples. The samples were rotated at about 0.5 rpm during sputtering. The sputtering process was terminated automatically by the thickness monitor at a coating thickness of approximately 2–3 nm.
Mechanical property testing
The mechanical tensile stress properties of the produced cellulose and biomaterials were measured using a tensile machine at a stretch rate of 0.4 mm s−1. The samples were cut to make uniform 2 × 1 mm pieces for testing. Samples were fixed between the upper and the lower clamps in order to prevent slip with force applied to the upper clamp to pull the sample in tension. The tensile experiments were performed at room temperature. The tensile stress σ* measured in Pa is defined as the force applied on the cellulose material (F) divided by the measured cross-sectional area of the sample (A) as shown in eqn (1)| σ* = F/A | (1) |
The tensile strain (ε) was calculated using eqn (2):
| ε = ΔL/L0 = (L − L0)/L0 | (2) |
| E = dσ*/dε | (3) |
Chemical analysis
Elemental composition (C, H, N) of the produced bacterial cellulose (BC) was determined by elemental analysis in a CHN Carlo Erba EA1108 CHN analyser. Films of BC microfibers were dried overnight at 70 °C to remove moisture before analysis. The elemental analysis was carried out after washing the cellulose several times with ethanol and deionised water in order to devoid the material of any cells or other media components. The bacterial cellulose and the produced biomaterial were also analysed using a FT-IR spectrophotometer. The samples were dried for 3 hours at 100 °C to remove moisture before the measurement of the FT-IR spectra. The spectra were recorded in the ATR mode on a Thermo Scientific Nicolet iS5 spectrophotometer in the range between 500 and 4000 cm−1 at a resolution of 4 cm−1 and a total accumulation of 64 scans. The samples were also examined using a Zeiss Evo 60 Scanning Electron Microscope equipped with an EDX energy dispersive X-ray analyser to investigate the elemental composition of the system (Inca energy 350) connected to an 80 mm2 silicon drift detector (SDD).Results and discussion
Growth media optimisation
The cell immobilisation procedure described in the Methods section was used to entrap the microalgae cells within the cellulose gel matrix produced by the acetobacter cells. The cells were grown in mixed media of various ratios of GY and TAP media with acetobacter and microalgae cells as shown in Scheme 1 and Fig. 2. Crucially important for the efficient growth and sustainability of both microorganisms in the system was to determine the optimal ratio of the GY and TAP media which also allows us to immobilise the microalgae efficiently in the cellulose gel produced by the acetobacter. We found that the volume ratio of 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 (v/v) GY
30 (v/v) GY![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
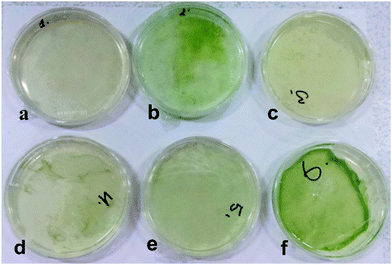
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TAP media is the optimal media formulation for efficient immobilisation of the microalgae within the bacterial cellulose matrix and the simultaneous growth of both microorganisms (see also Fig. 2f). A larger amount of GY media in the system compared to TAP media was needed as more glucose was necessary for the synthesis of bacterial cellulose. The viability of the immobilised C. reinhardtii cells in the cellulose gel produced by A. aceti was checked using FDA as a live/dead cell assay at different stages of growth of the living material. The viability of the immobilised cells was maintained for up to 7 days after which the biomaterial was extracted for further analysis. It is evident from Fig. 3 that the immobilised microalgae cells were viable under all media conditions with different ratios of GY and TAP media. However, we found large variations in the morphology and the texture of the produced living biomaterial. More results and analysis including SEM images of these materials are presented in the ESI.†
TAP media is the optimal media formulation for efficient immobilisation of the microalgae within the bacterial cellulose matrix and the simultaneous growth of both microorganisms (see also Fig. 2f). A larger amount of GY media in the system compared to TAP media was needed as more glucose was necessary for the synthesis of bacterial cellulose. The viability of the immobilised C. reinhardtii cells in the cellulose gel produced by A. aceti was checked using FDA as a live/dead cell assay at different stages of growth of the living material. The viability of the immobilised cells was maintained for up to 7 days after which the biomaterial was extracted for further analysis. It is evident from Fig. 3 that the immobilised microalgae cells were viable under all media conditions with different ratios of GY and TAP media. However, we found large variations in the morphology and the texture of the produced living biomaterial. More results and analysis including SEM images of these materials are presented in the ESI.†
We experimented with various amounts of A. aceti and a constant amount of C. reinhardtii in the 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 GY
30 GY![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
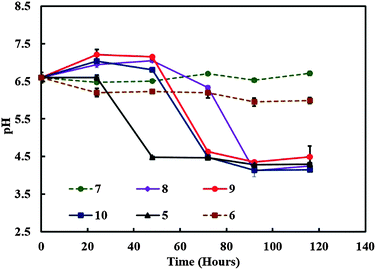
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TAP media (v/v) in the symbiotic culture vessel. The pH was recorded at various times in order to monitor the change in the culture media acidity which usually varies due to the accumulation of by-products of the cell metabolism. In samples containing both kinds of microorganisms the pH decreased strongly from the initial value of 6.5 as observed from Fig. 4. The pH of the control samples containing only A. aceti showed very little change from the initial pH value with time, while in the case of C. reinhardtii it dropped from the initial value of 6.5 to 4.5 as shown in Fig. 4. In another experiment we varied the amount of C. reinhardtii cells keeping the amount of A. aceti cells constant in the 70
TAP media (v/v) in the symbiotic culture vessel. The pH was recorded at various times in order to monitor the change in the culture media acidity which usually varies due to the accumulation of by-products of the cell metabolism. In samples containing both kinds of microorganisms the pH decreased strongly from the initial value of 6.5 as observed from Fig. 4. The pH of the control samples containing only A. aceti showed very little change from the initial pH value with time, while in the case of C. reinhardtii it dropped from the initial value of 6.5 to 4.5 as shown in Fig. 4. In another experiment we varied the amount of C. reinhardtii cells keeping the amount of A. aceti cells constant in the 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 GY
30 GY![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TAP media (v/v). It was observed that cultures with a substantial amount of C. reinhardtii growth showed a similar decrease in pH from 6.5 to 4.5 approximately as shown in Fig. 5. Drop in pH of the media was not observed in the case of cultures without microalgae growth. It can be concluded that under these conditions, the metabolism of the microalgae in the system increases the acidity of the surrounding culture medium. It is known that the microalgae can use the residual glucose present in the culture media in the process of glycolysis in order to produce additional energy (ATP) and release pyruvate (pyruvic acid) as a by-product.20 This explains the reduction in the pH with time as evident from Fig. 4 and 5 with the growth of the microalgae. The drop in the pH was highly significant and much quicker in the case of culture containing only C. reinhardtii compared to cultures containing both the microorganisms, which is due to the availability of ample glucose for the microalgae. Whereas the presence of acetobacter results in the use of glucose for its own metabolism.
TAP media (v/v). It was observed that cultures with a substantial amount of C. reinhardtii growth showed a similar decrease in pH from 6.5 to 4.5 approximately as shown in Fig. 5. Drop in pH of the media was not observed in the case of cultures without microalgae growth. It can be concluded that under these conditions, the metabolism of the microalgae in the system increases the acidity of the surrounding culture medium. It is known that the microalgae can use the residual glucose present in the culture media in the process of glycolysis in order to produce additional energy (ATP) and release pyruvate (pyruvic acid) as a by-product.20 This explains the reduction in the pH with time as evident from Fig. 4 and 5 with the growth of the microalgae. The drop in the pH was highly significant and much quicker in the case of culture containing only C. reinhardtii compared to cultures containing both the microorganisms, which is due to the availability of ample glucose for the microalgae. Whereas the presence of acetobacter results in the use of glucose for its own metabolism.
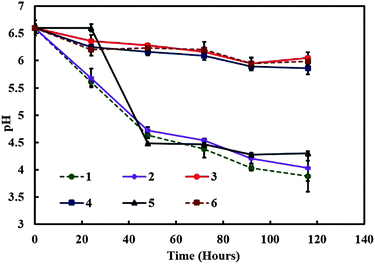 | ||
| Fig. 4 The media pH versus time for various mixed cultures containing different amounts of C. reinhardtii and A. aceti. The numbered lines represent the specific sample setup from Table 1. | ||
 | ||
| Fig. 5 The media pH versus time for various mixed cultures containing different amounts of C. reinhardtii and A. aceti. The numbers represent the specific sample setup from Table 2. | ||
These sets of experiments enabled us to pinpoint the specific amount to acetobacter and microalgae required in order to maintain successful symbiotic consortia and in turn control the production of the living biomaterial.
SEM studies of the living soft matter microstructure
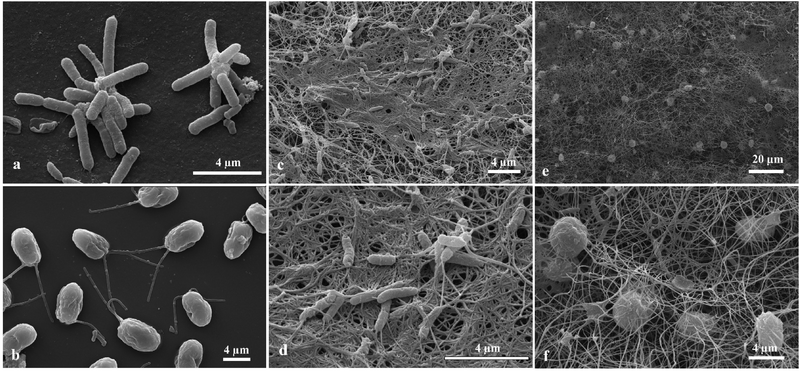
The microalgae C. reinhardtii mimic the photosynthetic alga and A. aceti mimics the filamentous fungi, which is the other part of the symbiotic pair in lichens. There are also some visual similarities in the morphology between lichens and the produced living biomaterials as presented in Fig. 6. The SEM image shown in Fig. 6a shows a natural lichen20 compared to the living biomaterial made in our laboratory as shown in Fig. 6b. The similarity in the microstructure of the immobilised microalgae in lichens compared to our living biomaterial is quite intriguing, although the latter is based on a completely different symbiotic pair and the extracellular matrix of bacterial cellulose nanofibers generated by A. aceti.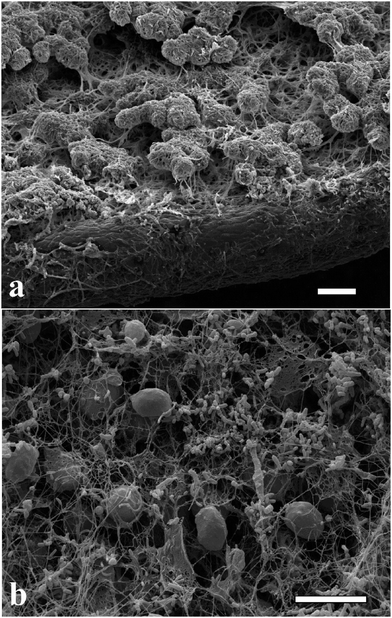 | ||
| Fig. 6 Scanning electron micrograph of (a) lichen colony growing on a leaf20 and (b) the living biomaterial made of cellulose produced from A. aceti and immobilised microalgae C. reinhardtii co-cultured in a glass beaker. Image (a) reproduced with permission from ref. 20. All scale bars are 10 μm in size. | ||
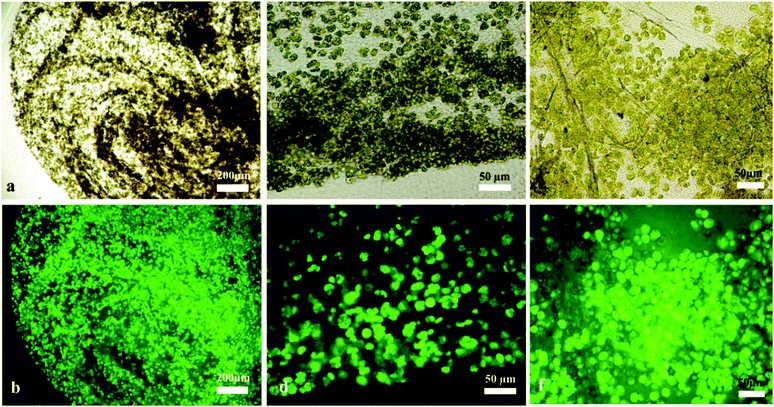
Samples from the living biomaterial composites were observed by SEM. The rod-shaped cells (Fig. 7a) and the oval shaped cells with two anterior flagella (Fig. 7b) are A. aceti and C. reinhardtii, respectively. When the cells were grown together in a Petri dish, the morphology of the produced biocomposite was different from the one produced in a beaker as shown in Fig. 7e, f and 8e, f. The number of cellulose-producing bacteria and microalgae cells are both much higher in the case of the biocomposites made in the beaker compared to those made in a Petri dish as evident from Fig. 7f and 8f. Note that the cellulose fibres are finer in the case of the thin biomaterial made in the Petri dish compared to the thick one in the beaker.
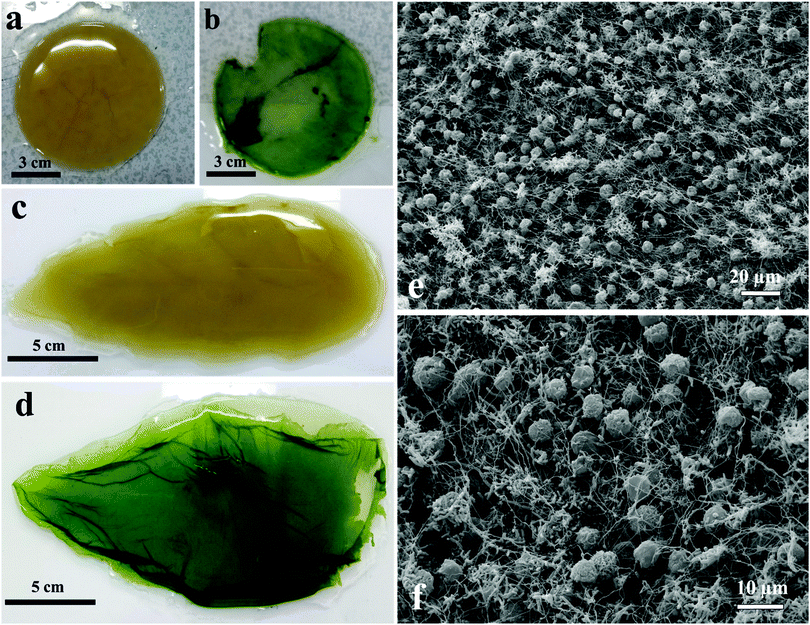
Moulding of the living biomaterial
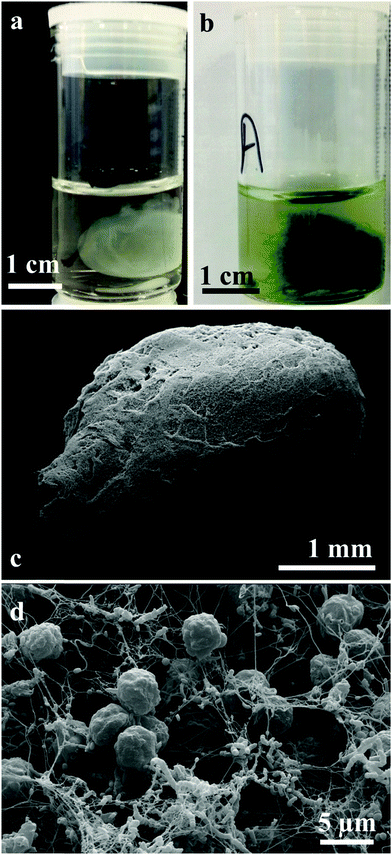
We have grown living biomaterial samples of different shapes like spherical bead, cylindrical and leaf-shaped. The shapes like flat circular thick cellulose as shown in Fig. 8a and b were made in a beaker while the leaf-shaped living biomaterial was produced in a culture vessel with the shape of a leaf as shown in Fig. 8c and d with the latter with immobilised microalgae in all cases. It was also observed that the cellulose sheet produced always takes the shape of the culture vessel, hence this property was used in order to “mold” the produced living soft matter into various shapes, sizes and thickness as shown in Fig. 8 and 11.We produced living soft matter in the shape of a spherical bead as shown in Fig. 11a and b where the latter was with immobilised C. reinhardtii cells grown during the cellulose bead formation. This was previously demonstrated by Hu et al.12 for the immobilisation of human osteoblast cells in sphere-like cellulose particles through the interactions between Gluconacetobacter xylinus and the mammalian cells. The SEM image analysis of the living soft matter bead presented in Fig. 11d shows similar morphological characteristics to the living biomaterial produced in the beaker – see Fig. 8f. In the ESI,† we also present different strategies for the integration of microalgae into a pre-fabricated slurry of bacterial cellulose gel beads with A. aceti.
Mechanical properties of the living biomaterial
We measured the elastic properties of the material using tensile strength equipment. The elastic modulus of the produced cellulose and the living biomaterial was found to be 510 ± 30 kPa and 120 ± 2 kPa respectively as shown in Fig. 9(A). The ultimate tensile strength of the materials was found to be 280 ± 10 kPa for the bacterial cellulose gel and 40 ± 2 kPa for the living biomaterial shown in Fig. 9(B). The calculations were based on the stress–strain graph shown in Fig. S10 (see ESI†). The bacterial cellulose gel material produced is much stronger and elastic in nature compared to the living biomaterial. Despite the lower mechanical strength compared with the original matrix the living biomaterial is strong enough to be handled and shaped in a variety of molds.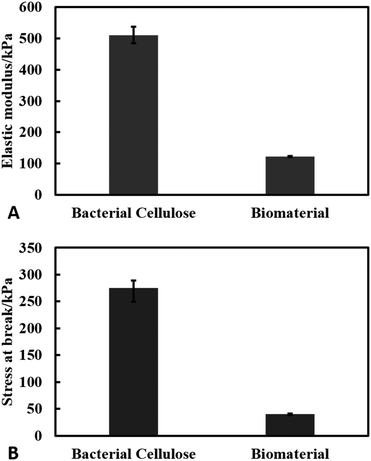 | ||
| Fig. 9 Results of tensile test of bacterial cellulose and the biomaterial. (A) Young's modulus (kPa) and (B) stress at break (kPa). The experiments were conducted in triplets. | ||
Characterisation of the biomaterial composition
The carbon and hydrogen contents of 45.30 ± 0.28 and 5.22 ± 0.26%, respectively, were found in dried samples of the symbiotic living biomaterial. These results were similar to the reported values of pure cellulose composition of carbon and hydrogen contents of 44.05 and 6.28%, respectively.21 Some nitrogen was also detected in the cellulose samples due to the presence of amino acids from the residual bacteria after purification.The FT-IR spectra of the produced bacterial cellulose as well as the living biomaterial are shown in Fig. 10. The samples obtained in the form of cellulose from A. aceti and the living biomaterial show similar bands to higher absorbance values in the case of the latter. The spectrum in Fig. 10 also shows the similar typical bands reported previously for cellulose based materials.22 The spectrum was also used to determine the ratio of allomorphs Iα and Iβ.23 The Iα/Iβ ratio was found to be 0.82 and 0.84 for cellulose and the living biomaterial respectively which is similar to that reported for bacterial cellulose.24 This also shows that the cellulose produced by A. aceti is rich in the Iα polymorph.
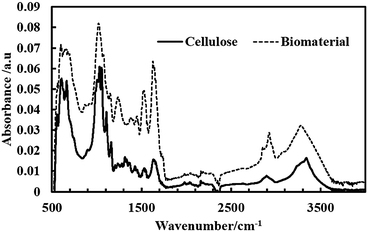 | ||
| Fig. 10 ATR FT-IR spectra of a film of bacterial cellulose produced by A. aceti and living biomaterial produced by the symbiotic growth of A. aceti and C. reinhardtii. | ||
The SEM-EDX analysis was conducted on the cellulose produced by A. aceti and also on the living biomaterials in order to determine the composition shown in Fig. S11 and S12 (ESI†). The results show higher percentages of carbon and oxygen as expected which correlated with our findings obtained using the CHN analyser. The presence of other elements like osmium, phosphorus, gold and palladium was observed due to sample preparation and coatings before SEM imaging and EDX analysis.
Conclusions
We have mimicked the relationship between microalgae and fungi, which forms lichens in Nature to produce a novel type of living soft matter based on the symbiosis of other unicellular microorganisms. It was found that A. aceti (NCIMB 8132), which can produce cellulose gels gets into a symbiotic relationship with the photosynthetic C. reinhardtii cells which produce oxygen. The microalgae cells were successfully integrated with the A. aceti cells in a symbiotic relationship by varying the concentration ratios of the initial culture media (TAP and GY media). It was found that at the ratio containing 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 GY
30 GY![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TAP media (v/v) both the microorganisms grow efficiently and integrate together within the produced bacterial cellulose gels. The immobilisation of the microalgae was done in real time along with the bacterial cellulose production. Since the microalgae C. reinhardtii have the ability to produce hydrogen under nutrient deprived conditions, such living soft matter may have advantages like improved hydrogen production13 and various other metabolite extractions. In the past, microalgae C. reinhardtii (cc124 strain) were immobilised in the alginate gel in order to increase their hydrogen production efficiency compared to suspension culture.13 The developed microalgae immobilisation procedure can be used to produce natural microalgae biofilms to be potentially integrated in the artificial leaf devices.13 The described approach of combining unicellular microorganisms working in symbiosis by passing metabolites to each other can lead to the development of other symbiotic pairs, where one type of cells builds the extracellular matrix in which the other type of cells integrates and provides essential metabolites. This technique may find applications in the development of a range of biological tissues and living soft matter systems and could be possibly extended to more than two symbiotic microorganisms, which might allow the creation of multifunctional, living and self-replicating systems.
TAP media (v/v) both the microorganisms grow efficiently and integrate together within the produced bacterial cellulose gels. The immobilisation of the microalgae was done in real time along with the bacterial cellulose production. Since the microalgae C. reinhardtii have the ability to produce hydrogen under nutrient deprived conditions, such living soft matter may have advantages like improved hydrogen production13 and various other metabolite extractions. In the past, microalgae C. reinhardtii (cc124 strain) were immobilised in the alginate gel in order to increase their hydrogen production efficiency compared to suspension culture.13 The developed microalgae immobilisation procedure can be used to produce natural microalgae biofilms to be potentially integrated in the artificial leaf devices.13 The described approach of combining unicellular microorganisms working in symbiosis by passing metabolites to each other can lead to the development of other symbiotic pairs, where one type of cells builds the extracellular matrix in which the other type of cells integrates and provides essential metabolites. This technique may find applications in the development of a range of biological tissues and living soft matter systems and could be possibly extended to more than two symbiotic microorganisms, which might allow the creation of multifunctional, living and self-replicating systems.
Acknowledgements
The authors are grateful to Mr Tony Sinclair for the SEM images and to Dr Tommy Horozov and Mr Khaled Althubeiti (University of Hull) who helped us with the tensile strength experiments. V. N. P acknowledges financial support from the EU COST Action CM1101.Notes and references
- S. D. Leavitt, E. Kraichak, M. P. Nelsen, S. Altermann, P. K. Divakar, D. Alors, T. L. Esslinger, A. Crespo and T. Lumbsch, Mol. Ecol., 2015, 24, 3779–3797 CrossRef PubMed.
- V. Ahmadjian, The lichen symbiosis, John Wiley and Sons Ltd, Chichester, England, 1993 Search PubMed.
- R. Honegger, Lichenologist, 1998, 30, 193–212 CrossRef.
- R. Honegger, New Phytol., 1993, 125, 659–677 CrossRef.
- M. Grube, M. Cardinale, J. V. de Castro, H. Muller and G. Berg, ISME J., 2009, 3, 1105–1115 CrossRef PubMed.
- B. P. Hodkinson and F. Lutzoni, Symbiosis, 2009, 49, 163–180 CrossRef CAS.
- T. D. Brock, M. T. Madigan, J. M. Martinko and J. Parker, Biology of microorganisms, Prentice-Hall, London, England, UK, 7th edn, 1994 Search PubMed.
- S. Jasti, M. E. Sieracki, N. J. Poulton, M. W. Giewat and J. N. Rooney-Varga, Appl. Environ. Microbiol., 2005, 71, 3483–3494 CrossRef CAS PubMed.
- Z. Guo and Y. Tong, J. Appl. Phycol., 2014, 26, 1483–1492 CrossRef CAS.
- N. M. N. Ton and V. V. M. Le, Int. Food Res. J., 2011, 18, 983 CAS.
- A. Rezaee, H. Godini and H. Bakhtou, Environ. Eng. Manage. J., 2008, 7, 589–594 CAS.
- Y. Hu, J. M. Catchmark and E. A. Vogler, Biomacromolecules, 2013, 14, 3444–3452 CrossRef CAS PubMed.
- A. A. K. Das, M. M. N. Esfahani, O. D. Velev, N. Pamme and V. N. Paunov, J. Mater. Chem. A, 2015, 3, 20698–20707 CAS.
- P. Raspor and D. Goranovic, Crit. Rev. Biotechnol., 2008, 28, 101–124 CrossRef CAS PubMed.
- R. A. Andersen, Algal culturing techniques, Elsevier/Academic Press, Burlington, Mass., 2005 Search PubMed.
- J. D. Fontana, A. M. De Souza, C. K. Fontana, I. L. Torriani, J. C. Moreschi, B. J. Gallotti, S. J. De Souza, G. P. Narcisco, J. A. Bichara and L. F. X. Farah, Appl. Biochem. Biotechnol., 1990, 24–25, 253–264 CrossRef CAS PubMed.
- T. H. Chrzanowski, R. D. Crotty, J. G. Hubbard and R. P. Welch, Microb. Ecol., 1984, 10, 179–185 CrossRef CAS PubMed.
- P. Breeuwer, J. Drocourt, F. M. Rombouts and T. Abee, Applied and Environmental Microbiology, 1996, 62, 178–183 CAS.
- P. Breeuwer, J. L. Drocourt, N. Bunschoten, M. H. Zwietering, F. M. Rombouts and T. Abee, Appl. Environ. Microbiol., 1995, 61, 1614–1619 CAS.
- L. L. Beer, E. S. Boyd, J. W. Peters and M. C. Posewitz, Curr. Opin. Biotechnol., 2009, 20, 264–271 CrossRef CAS PubMed.
- D. Klemm, D. Schumann, U. Udhardt and S. Marsch, Prog. Polym. Sci., 2001, 26, 1561–1603 CrossRef CAS.
- H. G. Shirk and G. A. Greathouse, Anal. Chem., 1952, 24, 1774–1775 CrossRef CAS.
- T. Imai and J. Sugiyama, Macromolecules, 1998, 31, 6275–6279 CrossRef CAS.
- C. Castro, R. Zuluaga, C. Álvarez, J.-L. Putaux, G. Caro, O. J. Rojas, I. Mondragon and P. Gañán, Carbohydr. Polym., 2012, 89, 1033–1037 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5tb02489g |
| This journal is © The Royal Society of Chemistry 2016 |