Prussian blue nanoparticles encapsulated inside a metal–organic framework via in situ growth as promising peroxidase mimetics for enzyme inhibitor screening†
Linjing
Su‡
,
Yuhao
Xiong‡
,
Haiguan
Yang
,
Peng
Zhang
and
Fanggui
Ye
*
Key Laboratory for the Chemistry and Molecular Engineering of Medicinal Resources (Ministry of Education of China), College of Chemistry and Pharmaceutical Science of Guangxi Normal University, Guilin 541004, P. R. China. E-mail: fangguiye@163.com; Fax: +86-773-5832294; Tel: +86-773-5856104
First published on 18th November 2015
Abstract
Artificial enzyme mimics are of current research interest owing to their remarkable advantages over natural enzymes. Herein, as a novel peroxidase mimic material, MIL-101(Cr)@PB was fabricated by encapsulating Prussian blue (PB) nanoparticles into the host matrix of MIL-101(Cr) via a facile and mild in situ growth synthetic strategy. The crystallographic characteristics, morphology, and porosity of the as-synthesized MIL-101(Cr)@PB composites were carefully studied using XRD, SEM, TEM, TGA, and BET. The results show that the synthesized MIL-101(Cr)@PB possesses a reproducible and impressive intrinsic peroxidase-like activity even under extreme conditions. Exploiting this, a colorimetric platform for screening xanthine oxidase inhibitors was constructed. We hope that this work will elucidate the applications of metal–organic frameworks as carriers for enzyme mimics and enable a wider application in drug screening.
Introduction
Over the last decade, nanomaterial-based artificial enzyme mimics have garnered increasing interest owing to their remarkable advantages over natural enzymes, such as their low cost, ease of storage, and good tunability for catalytic activity.1,2 Since Gao et al. discovered inert ferromagnetic nanoparticles with peroxidase-like activity, there has been a possibility for developing nanomaterials as nanozymes to mimic peroxidase activity. In the meantime, a great deal of excellent work has been done, including carbon-based nanomaterials (such as carbon nanotubes, graphene oxide, and graphitic carbon nitride nanosheets),3–5 noble-metal nanomaterials (such as gold and platinum nanoparticles),6–8 metal oxides (such as V2O5, Co3O4, WO3, and CeO2),9–12 and other composite materials.13–20 These excellent studies have promoted the development of artificial enzyme mimics; however, their inherent disadvantages are also obvious and include aggregation and settlement of the nanozymes in aqueous systems, which affects their catalytic activity and applications, causes difficulty in separation and impaired reusability, and leads to laborious preparation procedures.21,22 Therefore, the development of innovative enzyme mimics with an excellent stability and high catalytic activity remains of great interest.Prussian blue (PB), which is an important class of mixed-valence compounds, has been extensively explored for the construction of electrochemical biosensors because of its excellent electrochemical behavior.23–25 PB has a high peroxidase-like catalytic activity towards hydrogen peroxide;26 however, very little attention has been paid to directly utilizing PB as a peroxidase mimetic for building colorimetric sensor platforms.27–29 There are two main constraints to this application: firstly, the blue color of PB itself affects the target signals, and secondly, PB is not stable and is prone to aggregation in aqueous solutions, which might affect the catalytic activity. In addition, hydroxyl ions, which are generated during PB-catalyzed reduction of hydrogen peroxide, could destroy the bond between iron(III) and nitrogen, thereby suppressing the activity of PB.24 Therefore, PB could be a promising peroxidase mimetic for colorimetric sensing if these issues are solved.
Metal–organic frameworks (MOFs) are a new class of functional crystalline porous materials that have emerged over the past decade.30 Very recently, they have attracted considerable attention for the entrapment of different active functional guest species to enhance the stability and dispersion of nanozymes.31–35 Using MOFs as carriers of functional guest species provides several advantages; these include a large internal surface area, increased thermal and chemical stability, and, most importantly, ultra-high porosity that could facilitate the selective transport of small molecules through the porous coating of the MOFs to protect the active functional guest.32 For example, gold nanoparticles,36 platinum nanoparticles,31 porphyrin,37 hemin,34 and cytochrome c33 have been encapsulated into MOFs to construct sensing platforms. However, the encapsulation of PB by MOFs has not yet been explored. The resulting more stable and robust PB-based heterostructures generated using advanced interfacial assembly technologies are promising for clinical, medical, food safety, and environmental protection applications.38 Therefore, it is of high interest and importance to incorporate PB into MOFs to generate novel nanozymes and explore new potential applications.
Encouraged by this possibility, herein, we pursue the filling of active functional guest PB into the cavities of MIL-101(Cr) to produce MIL-101(Cr)@PB via a facile and mild in situ growth synthetic strategy. In addition, the excellent peroxidase-like catalytic activity of MIL-101(Cr)@PB is explored. Compared to bare PB, the synthesized MIL-101(Cr)@PB demonstrates several prominent advantages: (1) the blue interference of PB was reduced for colorimetric sensing; (2) the resistance to extreme conditions, such as extreme pH values and temperatures, was improved; and (3) the catalytic activity and synthesis of MIL-101(Cr)@PB were reproducible, and the catalyst was reusable. Based on the excellent peroxidase-like catalytic activity of MIL-101(Cr)@PB, an inexpensive, simple, and rapid colorimetric platform for screening xanthine oxidase (XO) inhibitors was constructed.
Materials and methods
Cr(NO3)3·9H2O, terephthalic acid (H2BDC), and N,N-dimethylformamide (DMF) were purchased from Sinopharm (Shanghai, China). Hydrofluoric acid (HF), ammonium fluoride (NH4F), FeCl2·4H2O, and kaempferol were purchased from Aladdin Chemistry (Shanghai, China). Potassium ferrocyanide (K4[Fe(CN)6]·3H2O) and H2O2 (30 wt%) were purchased from Shanghai Chemical Reagents Corporation (Shanghai, China). 3,3′,5,5′-Tetramethylbenzidine (TMB) was purchased from TCI (Shanghai, China). Xanthine oxidase (10 U mL−1) was purchased from Sigma-Aldrich (Shanghai, China), and xanthine was purchased from J&K Chemical Technology (Beijing, China).Powder X-ray diffraction (XRD) patterns were performed on a D/max 2550 VB/PC diffractometer (Rigaku, Japan). Fourier transform infrared (FT-IR) spectra (4000–400 cm−1) were recorded in KBr discs on a PE Spectrum One FT-IR spectrometer (Perkin Elmer, USA). Thermogravimetric analysis (TGA) was performed within the temperature range of 30–800 °C on a LABSYS Evo TGDSC/DTA instrument (Setaram Instrumentation, France). Scanning electron microscopy (SEM) was carried out on an FEI Quanta 200 FEG scanning electron microscope (Philips, The Netherlands). Transmission electron microscopy (TEM) images were obtained on an FEI TECNAI G20 instrument (FEI, USA). Nitrogen adsorption and desorption at 77 K was performed using a 3Flex adsorption instrument (Micromeritics, USA) to evaluate the pore structures, pore size distributions, and pore volumes via the Barrett–Joyner–Halenda (BJH) method. UV-Vis absorption spectra were recorded on a Cary 60 spectrophotometer (Agilent, USA). Ultrapure water was produced using a Millipore purification system (Bedford, MA, USA) and used to prepare all aqueous solutions.
Synthesis and purification of MIL-101(Cr)
MIL-101(Cr) was synthesized according to the report by Férey et al., as follows:39,40 800 mg of Cr(NO3)3·9H2O (2.0 mmol), 332 mg of H2BDC (2.0 mmol), and 0.1 mL of HF (2.0 mmol) were mixed with 9.6 mL of ultrapure water in a Teflon-lined bomb, then reacted in an oven and heated at 220 °C for 8 h. The obtained green solid was collected by filtration and washed five times with ethanol. The as-synthesized MIL-101(Cr) was further purified using hot ethanol and aqueous NH4F solutions. Typically, MIL-101(Cr) was dispersed in water at 70 °C for 5 h, refluxed in hot ethanol at 60 °C for 3 h, and then dispersed in 150 mL of an aqueous solution of 30 mM NH4F at 60 °C for 10 h; after each step, the product was collected by filtration. Finally, the precipitate was washed five times with 200 mL of hot water (60 °C) to remove any traces of NH4F and dried at 150 °C in vacuo for 12 h to form dehydrated MIL-101(Cr).Synthesis of MIL-101(Cr)@PB
MIL-101(Cr)@PB was synthesized using a facile and mild in situ growth method. In particular, 100 mg of the as-synthesized MIL-101(Cr) was mixed with 40 mL of a DMF solution containing K4[Fe(CN)6] (0.4 mM) and the mixture underwent vigorous magnetic stirring for 3 h at room temperature. Subsequently, the precipitate was collected by centrifugation, washed once with ethanol, and then dispersed in 20 mL of ultrapure water to form a suspension. Then, 20 mL of aqueous solution containing FeCl2 (1.0 mM) was added to the suspension, which then underwent vigorous magnetic stirring for 3 h at room temperature. Finally, the light blue composites were collected by centrifugation, washed several times with ethanol, and dried at 60 °C in vacuo for 12 h. A spectrophotometric method was used to determine the PB content in MIL-101(Cr) (the details are shown in the ESI†).Peroxidase-like activity measurements
The peroxidase-like activity of MIL-101(Cr)@PB was investigated via catalytic oxidation of a peroxidase substrate, TMB, in the presence of H2O2. The solutions used for the kinetic analyses with TMB as the substrate consisted of 750 μL of NaAc buffer (0.1 M pH 4.0), 50 μL of MIL-101(Cr)@PB solution (2.0 mg mL−1), 100 μL of 9.8 mM H2O2, and 100 μL of various concentrations of TMB (i.e., 0, 0.2, 0.5, 0.8, 1.1, 1.4, 1.7, and 2.0 mM). Similarly, the solutions used for the kinetic analysis of H2O2 as the substrate consisted of 750 μL of NaAc buffer (0.1 M pH 4.0), 50 μL of MIL-101(Cr)@PB solution (2.0 mg mL−1), 100 μL of 1.4 mM TMB, and 100 μL of varying concentrations of H2O2 (i.e., 0, 0.49, 0.98, 1.96, 3.92, 5.88, 7.84, and 9.8 mM). For the experiment of PB NPs, the detection procedure was the same as that of MIL-101(Cr)@PB but using PB NPs (0.2 mg mL−1, the same as the amount of PB in MIL-101(Cr)@PB) instead of MIL-101(Cr)@PB. The kinetic parameters were calculated based on Lineweaver–Burk plots of the double reciprocal of the Michaelis–Menten equation, as follows:| 1/V = Km/Vmax(1/[C] + 1/Km), | (1) |
Solvent extraction of Selaginella pulvinata
Selaginella pulvinata was purchased from a local Chinese herbal medicine market. After being air-dried, Selaginella pulvinata was crushed in a mill and a 0.5 g sample was dispersed in a 50% v/v methanol/water solution (or 100% water) at 80 °C in a water bath for 2.0 h. Then, after filtering and collecting the filtrate, it was evaporated to near dryness using vacuum distillation. The final extract was diluted to 50 mL using ultrapure water and stored in a refrigerator at 4 °C.Screening of xanthine oxidase inhibitors
First, 20 μL of XO solution (0.15 U mL−1), 80 μL of xanthine (1.0 mM), and 50 μL of inhibitor at different concentrations were mixed and incubated for 1 h at room temperature; then 300 μL of acetate buffer (0.1 M, pH 4.0), 20 μL of TMB ethanol solution (10 mM), and 20 μL of MIL-101(Cr)@PB solution (2.0 mg mL−1) were successively added to the xanthine reaction solution, which was then incubated for 15 min at 37 °C; finally, the mixed reaction solution was filtered through a 0.25 μm nylon membrane filter to remove MIL-101(Cr)@PB. The absorbance at 655 nm was then measured, and the inhibition was calculated using the following equation:| Inhibition ratio (%) = 100[(A0 − A)/A0], | (2) |
Results and discussion
Synthesis and characterization of MIL-101(Cr)@PB
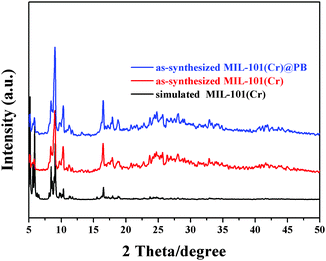
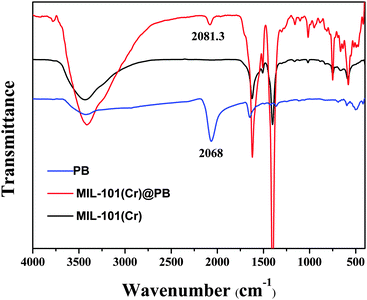
The preparation of MIL-101(Cr)@PB was carried out using a facile and mild in situ growth synthetic strategy. The Fe(CN)63− ions are able to completely enter the pores of MIL-101(Cr) because the pore windows (pentagonal window: ∼12 Å, hexagonal window: ∼16 Å) are larger than a Fe(CN)63− ion (hydrated diameter: ∼9.4 Å); also, MIL-101(Cr) adsorbs Fe(CN)63− easily and steadily because of anion exchange between Fe(CN)63− and the OH−/F− ions attached to the Cr metal site of MIL-101 (Fig. S1, ESI†).39,41 Then, removing the free Fe(CN)63− from the outer surface of the MIL-101(Cr) by washing and adding Fe2+ solution resulted in the formation of MIL-101(Cr)@PB. The crystalline structures of MIL-101(Cr) and MIL-101(Cr)@PB were analyzed using a powder XRD technique (Fig. 1); the XRD patterns of the as-synthesized MIL-101(Cr) and a simulated structure were in excellent agreement, indicating that MIL-101(Cr) was successfully prepared. For MIL-101(Cr)@PB, the pattern showed the same reflections as the pure MIL-101(Cr) sample, which illustrates that the encapsulation of PB did not disrupt the crystal structure of MIL-101(Cr). However, no diffraction peaks from PB crystals were detected; this is likely because of the low content and the small size of the PB NPs. As shown in Fig. 2, a characteristic absorbance band at 2068 cm−1 was observed for PB and attributed to the C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N stretching vibration. However, this absorbance band for MIL-101(Cr)@PB was blue-shifted by 13.3 cm−1, which indicates an interaction between MIL-101(Cr) and PB.42
N stretching vibration. However, this absorbance band for MIL-101(Cr)@PB was blue-shifted by 13.3 cm−1, which indicates an interaction between MIL-101(Cr) and PB.42
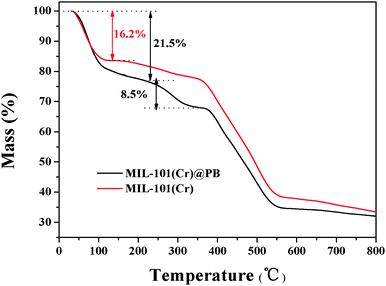
The content of PB in MIL-101(Cr) was evaluated by TGA analysis (Fig. 3) and spectrophotometric methods (Fig. S2, ESI†). The first main weight-loss stage was due to the dehydration of the material up to 110 °C. It is clear from Fig. 3 that the water content in MIL-101(Cr)@PB increased as the amount of PB increased; this is because PB contains water molecules. The second weight loss observed at about 250 °C mainly indicates the decomposition of PB. According to the TGA analysis and spectrophotometric method results, the calculated amount of PB in MIL-101(Cr)@PB was about 8.5%.
The morphology and the microstructure of MIL-101(Cr)@PB were characterized in detail using SEM and TEM (Fig. 4 and Fig. S3, ESI†). The SEM images (Fig. 4A and B) clearly showed that both MIL-101(Cr)@PB and MIL-101(Cr) have essentially the same morphology and are defined as octahedral crystals with average diameters of less than 500 nm; this result also shows that in situ growth of PB did not disrupt the crystal structure of MIL-101(Cr). The TEM images show that PB NPs were uniformly distributed in the MIL-101(Cr) matrix (Fig. 4C and D and Fig. S3A, ESI†). The representative high-resolution transmission electron microscopy (HRTEM) images (Fig. S2B–D, ESI†) show that the average size of the PB NPs is ∼2 nm, and typical single PB nanocrystals with a highly crystalline texture were observed. The HRTEM images also indicate that the lack of PB peaks in the MIL-101(Cr)@PB XRD patterns was not caused by the amorphous nature of the PB NPs in MIL-101(Cr); this result differs from that in a previous study.42
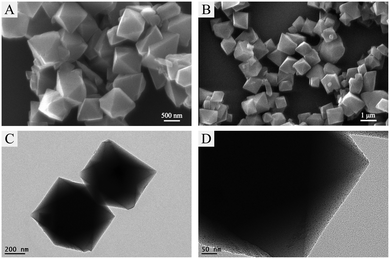 | ||
| Fig. 4 SEM images of (A) MIL-101(Cr) and (B) MIL-101(Cr)@PB and TEM images of (C and D) MIL-101(Cr)@PB. | ||
The surface area is one of the most significant factors that influence the catalytic activity of catalysts. As shown in Table 1 and Fig. S4 and S5 (ESI†), the decrease in the amount of nitrogen adsorption and pore volume of MIL-101(Cr)@PB reveal that the cavities of MIL-101(Cr) were partially occupied by highly dispersed PB NPs; nevertheless, the BET surface area remained as high as 1235.1 m2 g−1. Thus, the high surface areas of MIL-101(Cr)@PB may provide more active centers and improve the catalytic capability. Considering the results of the FT-IR, SEM, TEM, XRD analyses, etc., we can conclude that MIL-101(Cr)@PB was successfully synthesized.
| Samples | BET surface area (m2 g−1) | BJH adsorption average pore width (nm) | BJH desorption average pore width (nm) | Pore volumes (cm3 g−1) |
|---|---|---|---|---|
| MIL-101(Cr) | 2711.9 | 2.21 | 2.19 | 1.4 |
| MIL-101(Cr)@PB | 1235.1 | 2.03 | 2.03 | 0.66 |
Peroxidase-like activity of MIL-101(Cr)@PB
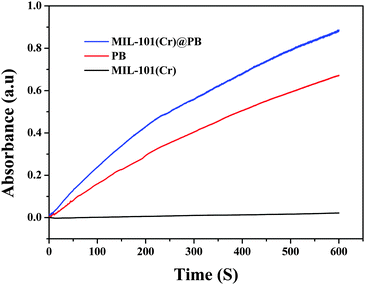
The peroxidase-like activity of MIL-101(Cr)@PB was evaluated via catalytic oxidation of TMB and other peroxidase substrates in the presence of H2O2. As shown in Fig. 5, no obvious oxidative reaction occurred with only the addition of H2O2 or MIL-101(Cr)@PB to the TMB solution. In contrast, the TMB–H2O2 system in the presence of MIL-101(Cr)@PB exhibited a strong absorbance at 655 nm. In addition, the time-dependent absorbance changes at 655 nm (Fig. 6) and UV-vis spectra (Fig. S6, ESI†) of MIL-101(Cr), PB, or MIL-101(Cr)@PB showed that MIL-101(Cr) did not have peroxidase-like activity. However, MIL-101(Cr)@PB exhibited much higher activity than bare PB, which suggests that the MIL-101(Cr) matrix improved the peroxidase-like activity of PB. To further characterize the peroxidase-like activity of MIL-101(Cr)@PB, several other typical peroxidase substrates were investigated (Fig. S7, ESI†). All the results indicate that MIL-101(Cr)@PB possesses remarkable peroxidase-like activity. In addition, Fig. S8 (ESI†) indicates that the absorbance change greatly increased with increasing MIL-101(Cr)@PB concentration and reached a maximum value at 2.0 mg mL−1. To better understand the peroxidase-like activity of MIL-101(Cr)@PB, steady-state kinetic parameters for the reaction in TMB and H2O2 were determined. TMB and H2O2 concentration-dependent reaction rate curves are shown in Fig. S9 (ESI†). The curves display typical Michaelis–Menten behavior. The Michaelis–Menten constant (Km) and maximum initial velocity (Vmax) were obtained using a Lineweaver–Burk plot (Table 2). Generally, Km characterizes the binding affinity of the enzyme for the substrates and a lower Km value implies a stronger affinity. These parameters show that the synthesized MIL-101(Cr)@PB possessed a higher affinity for TMB and H2O2 than HRP and other nanozymes (Table S1, ESI†).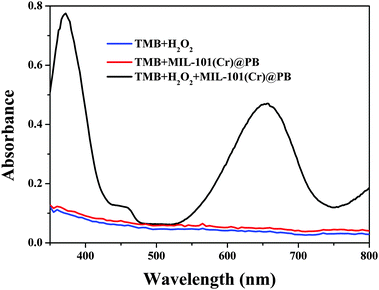 | ||
| Fig. 5 UV-Vis spectra of a TMB buffer solution containing H2O2, MIL-101(Cr)@PB, and MIL-101(Cr)@PB/H2O2. | ||
| Catalyst | Substance | K m/mM | V max/10−8 M s−1 | K cat/s−1 |
|---|---|---|---|---|
| MIL-101(Cr)@PB | TMB | 0.88 | 11.5 | 9.9 × 102 |
| H2O2 | 1.06 | 9.7 | 8.4 × 102 | |
| PB | TMB | 0.91 | 2.7 | 2.3 × 102 |
| H2O2 | 7.82 | 2.4 | 2.1 × 102 |
Good reusability is essential for an excellent catalyst; therefore, the reusability of MIL-101(Cr)@PB was investigated by conducting the experiment independently over five cycles. As shown in Fig. S10 (ESI†), the crystalline structures and morphology of MIL-101(Cr)@PB did not change after five cycles, and the results reveal no observable loss of activity. These data indicate the extraordinary stability of MIL-101(Cr)@PB and its excellent reproducibility.
Comparison of the peroxidase-like activity of MIL-101(Cr)@PB and bare PB under different conditions
It is well-known that PB has poor stability in alkaline solutions resulting in decreased activity. Therefore, in order to evaluate the stability of MIL-101(Cr)@PB towards alkaline degradation, we soaked MIL-101(Cr)@PB in different alkaline solutions to investigate whether it affects the peroxidase-like activity. As shown in Fig. S11 (ESI†), the results revealed no loss of activity even up to pH 11. A further increase in the pH leads to the decomposition of MIL-101(Cr) and a sharp decrease in activity. However, Fig. S12 (ESI†) shows that the characteristic peak of PB at 2081.3 cm−1 disappeared and new peaks at about 2958 cm−1 appeared. The new peaks were assigned to CH3 asymmetric and symmetric stretching vibrations from carbon deposits.43 These data suggest that the remaining peroxidase-like activity of MIL-101(Cr)@PB after soaking in alkaline solutions was not from PB but rather from the generated iron hydroxo species. Although MIL-101(Cr) could not protect PB from alkali attack, it protected the iron hydroxo species that formed during PB decomposition. Thus, even after extended soaking periods in strong alkaline solutions, MIL-101(Cr)@PB retained excellent catalytic activity.Usually, natural enzymes readily deactivate at high temperatures. Unfortunately, the catalytic activity of bare PB also sharply decreases at high temperatures. As shown in Fig. S13 (ESI†), after harsh treatment of hemoglobin, bare PB, and MIL-101(Cr)@PB at 95 °C for 40 min, only ∼25% of the peroxidase-like activity of hemoglobin and bare PB remained. However, the peroxidase-like activity of MIL-101(Cr)@PB barely changed. These results clearly indicate the excellent preservation of the peroxidase-like activity of MIL-101(Cr)@PB after exposure to harsh heat treatment.
During the catalytic oxidation of TMB in the presence of H2O2, we found that bare PB was unstable and prone to aggregation (Fig. S14, ESI†). This may be because the surfaces of PB NPs are negatively charged and oxTMB is positively charged; therefore, electrostatic interactions cause PB to aggregate. However, the MIL-101(Cr) matrix could protect the PB NPs enabling MIL-101(Cr)@PB to be stable in catalytic systems. In addition, some impurities in the samples, such as proteins, also adsorb nanocatalysts and reduce their catalytic activity. As shown in Fig. S15 (ESI†), when model protein BSA was added, the peroxidase-like activity of bare PB decreased to ∼85% (Fig. S15a, ESI†). In contrast, no significant changes in peroxidase-like performance emerged for MIL-101(Cr)@PB (Fig. S15b, ESI†); this may be due to the ultra-high porosity of MOF, which could facilitate the selective transport of small molecules through the MIL-101(Cr) porous coating and protect the active functional guest PB NPs.
As mentioned in the Introduction, the dark blue color of PB interferes with the signals for colorimetric sensing. This interference is reduced for MIL-101(Cr)@PB because it is light blue. More importantly, MIL-101(Cr)@PB can be easily removed using low-speed centrifugation (<5000 rpm) or simple filtration. In contrast, bare PB NPs are very difficult to separate.
Screening of xanthine oxidase inhibitors using MIL-101(Cr)@PB
Since the catalytic activity of MIL-101(Cr)@PB is proportional to the H2O2 concentration and H2O2 is the main product of the XO-xanthine catalyzed reaction, a colorimetric method for screening XO inhibitors could be realized using MIL-101(Cr)@PB. As illustrated in Scheme 1, XO oxidizes xanthine to quantitatively form uric acid and H2O2. The resultant H2O2 is then utilized by MIL-101(Cr)@PB to oxidize TMB to produce a blue solution, which can be simply detected by the naked eye or using a UV-vis spectrometer. However, in the presence of an inhibitor, the XO activity was inhibited and the amount of H2O2 generated decreased correspondingly. Thus, we can achieve a simple and rapid XO-inhibitor screening by observing changes in the color of the solution.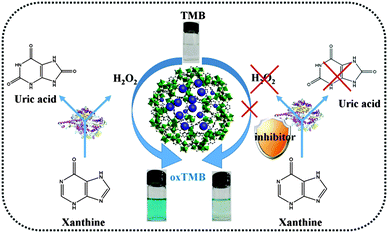 | ||
| Scheme 1 Schematic illustration of the colorimetric platform for screening of xanthine oxidase inhibitors. | ||
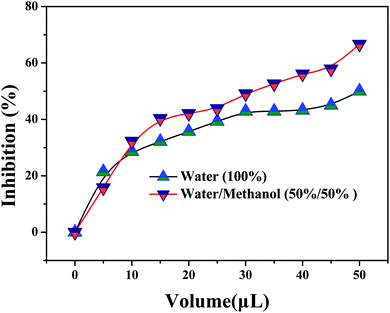
To prove this principle, kaempferol was used as an XO inhibitor.44 As shown in Fig. 7, with increasing kaempferol concentration, the absorbance gradually decreased. This means that the activity of XO was inhibited, which reduced the amount of H2O2 generated and lightened the blue color of the system (Fig. 7 inset). As shown in Fig. S16 (ESI†), at a kaempferol concentration of 46.58 μM, the inhibition ratio was nearly 85%. According to calculations, the IC50 was about 2.39 μM, which is very close to the reported values.44 In addition, a traditional Chinese herbal medicine, i.e., Selaginella pulvinata, was used as a real sample model for screening XO inhibitors. As shown in Fig. 8, with the increasing amount of extract, the XO activity inhibition ratio increased. Also, the inhibiting ability of Selaginella pulvinata extracted from a 50% v/v methanol/water solution was higher than that extracted using 100% water; this may be because the active ingredients in Selaginella pulvinata are non-polar and much more easily extracted by methanol. These results show that the proposed method is rapid and simple and could potentially be used for the initial screening of natural complex mixtures for XO inhibition.
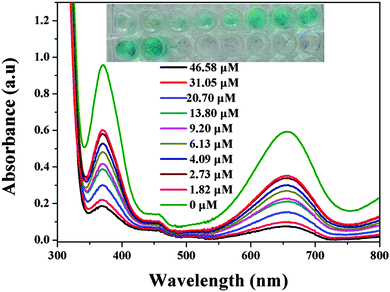 | ||
| Fig. 7 UV-Vis spectra of the XO reaction system in the presence of inhibitor kaempferol at different concentrations. Inset: corresponding photographs. | ||
Conclusions
In summary, MIL-101(Cr)@PB was fabricated by encapsulating PB NPs into the host matrix of MIL-101(Cr) using a facile and mild in situ growth synthetic strategy. The results showed that MIL-101(Cr)@PB possesses remarkable intrinsic peroxidase-like activity. More importantly, owing to the beneficial features of MIL-101(Cr), such as its protection of the catalytic capacity of PB NPs and enabling of the rapid diffusion of small molecules into the nanocages, MIL-101(Cr)@PB showed more efficient catalysis than PB NPs and higher stability in a broad range of conditions, such as high temperatures and aggregation. Accordingly, a colorimetric method of screening xanthine oxidase inhibitors was developed. Additionally, as a mimic peroxidase and compared to natural enzymes, MIL-101(Cr)@PB exhibited several advantages, such as easy preparation and preservation, low cost, and stability. Thus, we hope that MIL-101(Cr)@PB will be applied in a wide range of fields such as catalysis, environmental remediation, and drug-screening.Acknowledgements
The financial support of the National Natural Science Foundation of China (21365005), Guangxi Natural Science Foundation of China (2014GXNSFGA118002), Key Laboratory for the Chemistry and Molecular Engineering of Medicinal Resources (Guangxi Normal University), Ministry of Education of China (CMEMR2014-A09), and Innovation Project of Guangxi Graduate Education (YCSZ2015090) is gratefully acknowledged.Notes and references
- Y. Lin, J. Ren and X. Qu, Acc. Chem. Res., 2014, 47, 1097–1105 CrossRef CAS PubMed.
- H. Wei and E. Wang, Chem. Soc. Rev., 2013, 42, 6060–6093 RSC.
- Y. Song, K. Qu, C. Zhao, J. Ren and X. Qu, Adv. Mater., 2010, 22, 2206–2210 CrossRef CAS PubMed.
- J. Tian, Q. Liu, A. M. Asiri, A. H. Qusti, A. O. Al-Youbi and X. Sun, Nanoscale, 2013, 5, 11604–11609 RSC.
- J. Qian, X. Yang, Z. Yang, G. Zhu, H. Mao and K. Wang, J. Mater. Chem. B, 2015, 3, 1624–1632 RSC.
- F. Wang, X. Liu, C. Lu and I. Willner, ACS Nano, 2013, 7, 7278–7286 CrossRef CAS PubMed.
- Y. Jv, B. Li and R. Cao, Chem. Commun., 2010, 46, 8017–8019 RSC.
- G. Wu, S. He, H. Peng, H. Deng, A. Liu, X. Lin, X. Xia and W. Chen, Anal. Chem., 2014, 86, 10955–10960 CrossRef CAS PubMed.
- F. Natalio, R. André, A. F. Hartog, B. Stoll, K. P. Jochum, R. Wever and W. Tremel, Nat. Nanotechnol., 2012, 7, 530–535 CrossRef CAS PubMed.
- J. Yin, H. Cao and Y. Lu, J. Mater. Chem., 2012, 22, 527–534 RSC.
- Y. Ma, M. Zhao, B. Cai, W. Wang, Z. Ye and J. Huang, Chem. Commun., 2014, 50, 11135–11138 RSC.
- Z. Tian, J. Li, Z. Zhang, W. Gao, X. Zhou and Y. Qu, Biomaterials, 2015, 59, 116–124 CrossRef CAS PubMed.
- X. Sun, S. Guo, C. Chung, W. Zhu and S. Sun, Adv. Mater., 2013, 25, 132–136 CrossRef CAS PubMed.
- L. Wan, J. Liu and X. Huang, Chem. Commun., 2014, 50, 13589–13591 RSC.
- Y. Jiang, W. Wang, X. Li, X. Wang, J. Zhou and X. Mu, ACS Appl. Mater. Interfaces, 2013, 5, 1913–1916 CAS.
- Y. Wang, X. Zhang, Z. Luo, X. Huang, C. Tan, H. Li, B. Zheng, B. Li, Y. Huang, J. Yang, Y. Zong, Y. Ying and H. Zhang, Nanoscale, 2014, 6, 12340–12344 RSC.
- L. Su, J. Feng, X. Zhou, C. Ren, H. Li and X. Chen, Anal. Chem., 2012, 84, 5753–5758 CrossRef CAS PubMed.
- H. Zhao, Y. Dong, P. Jiang, G. Wang and J. Zhang, ACS Appl. Mater. Interfaces, 2015, 7, 6451–6461 CAS.
- Q. An, C. Sun, D. Li, K. Xu, J. Guo and C. Wang, ACS Appl. Mater. Interfaces, 2013, 5, 13248–13257 CAS.
- W. Gao, J. Li, X. Zhou, Z. Zhang, Y. Ma and Y. Qu, J. Mater. Chem. C, 2014, 2, 8729–8735 RSC.
- H. Tan, C. Ma, L. Gao, Q. Li, Y. Song, F. Xu, T. Wang and L. Wang, Chem. – Eur. J., 2014, 20, 16377–16383 CrossRef CAS PubMed.
- T. Zhang, Y. Lu and G. Luo, ACS Appl. Mater. Interfaces, 2014, 6, 14433–14438 CAS.
- L. Zhang and Y. Liu, Anal. Chim. Acta, 2014, 806, 204–209 CrossRef CAS PubMed.
- A. Michopoulos, A. Kouloumpis, D. Gournis and M. I. Prodromidis, Electrochim. Acta, 2014, 146, 477–484 CrossRef CAS.
- P. C. Pandey and A. K. Pandey, Electrochim. Acta, 2014, 125, 465–472 CrossRef CAS.
- H. Zhang, Y. Yin, P. Wu and C. Cai, Biosens. Bioelectron., 2012, 31, 244–250 CrossRef CAS PubMed.
- A. K. Dutta, S. K. Maji, P. Biswas and B. Adhikary, Sens. Actuators, B, 2013, 177, 676–683 CrossRef CAS.
- W. Zhang, D. Ma and J. Du, Talanta, 2014, 120, 362–367 CrossRef CAS PubMed.
- T. Wang, Y. Fu, L. Chai, L. Chao, L. Bu, Y. Meng, C. Chen, M. Ma, Q. Xie and S. Yao, Chem. – Eur. J., 2014, 20, 2623–2630 CrossRef CAS PubMed.
- H. Furukawa, K. E. Cordova, M. O'Keeffe and O. M. Yaghi, Science, 2013, 341, 1230444 CrossRef PubMed.
- Z. Xu, L. Yang and C. Xu, Anal. Chem., 2015, 87, 3438–3444 CrossRef CAS PubMed.
- K. Liang, R. Ricco, C. M. Doherty, M. J. Styles, S. Bell, N. Kirby, S. Mudie, D. Haylock, A. J. Hill, C. J. Doonan and P. Falcaro, Nat. Commun., 2015, 6, 7240–7246 CrossRef CAS PubMed.
- F. Lyu, Y. Zhang, R. N. Zare, J. Ge and Z. Liu, Nano Lett., 2014, 14, 5761–5765 CrossRef CAS PubMed.
- F. Luo, Y. Lin, L. Zheng, X. Lin and Y. Chi, ACS Appl. Mater. Interfaces, 2015, 7, 11322–11329 CAS.
- X. Wu, J. Ge, C. Yang, M. Hou and Z. Liu, Chem. Commun., 2015, 51, 13408–13411 RSC.
- Y. Hu, J. Liao, D. Wang and G. Li, Anal. Chem., 2014, 86, 3955–3963 CrossRef CAS PubMed.
- P. Ling, J. Lei, L. Zhang and H. Ju, Anal. Chem., 2015, 87, 3957–3963 CrossRef CAS PubMed.
- B. Kong, C. Selomulya, G. Zheng and D. Zhao, Chem. Soc. Rev., 2015, 44, 7997–8018 RSC.
- G. Férey, C. Mellot-Draznieks, C. Serre, F. Millange, J. Dutour, S. Surblé and I. Margiolaki, Science, 2005, 309, 2040–2042 CrossRef PubMed.
- D. Hong, Y. K. Hwang, C. Serre, G. Férey and J. Chang, Adv. Funct. Mater., 2009, 19, 1537–1552 CrossRef CAS.
- L. Gao, C. V. Li and K. Chan, Chem. Mater., 2015, 27, 3601–3608 CrossRef CAS.
- G. Liang, L. Zheng, S. Bao, H. Gao, F. Zhu and Q. Wu, Carbon, 2015, 93, 719–730 CrossRef CAS.
- I. O. Ali, T. M. Salama, M. S. Thabet, K. S. El-Nasser and A. M. Hassan, Mater. Chem. Phys., 2013, 140, 81–88 CrossRef CAS.
- Y. Wang, G. Zhang, J. Pan and D. Gong, J. Agric. Food Chem., 2015, 63, 526–534 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5tb01924a |
| ‡ These two authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2016 |