Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Optically assembled droplet interface bilayer (OptiDIB) networks from cell-sized microdroplets†
Mark S.
Friddin
 ab,
Guido
Bolognesi
ab,
Guido
Bolognesi
 a,
Yuval
Elani
ab,
Nicholas J.
Brooks
ab,
Robert V.
Law
ab,
John M.
Seddon
ab,
Mark A. A.
Neil
bc and
Oscar
Ces
*ab
a,
Yuval
Elani
ab,
Nicholas J.
Brooks
ab,
Robert V.
Law
ab,
John M.
Seddon
ab,
Mark A. A.
Neil
bc and
Oscar
Ces
*ab
aDepartment of Chemistry, Imperial College London, Exhibition Road South Kensington, London, SW7 2AZ, UK. E-mail: o.ces@imperial.ac.uk
bInstitute of Chemical Biology, Imperial College London, Exhibition Road South Kensington, London, SW7 2AZ, UK
cPhotonics Group, Department of Physics, Imperial College London, Exhibition Road South Kensington, London, SW7 2AZ, UK
First published on 12th September 2016
Abstract
We report a new platform technology to systematically assemble droplet interface bilayer (DIB) networks in user-defined 3D architectures from cell-sized droplets using optical tweezers. Our OptiDIB platform is the first demonstration of optical trapping to precisely construct 3D DIB networks, paving the way for the development of a new generation of modular bio-systems.
Droplet interface bilayers (DIBs) have recently emerged as a powerful and versatile tool for assembling a lipid bilayer between two aqueous microdroplets immersed in oil. The technique works by supplying lipids to either the water droplets (lipid-in) or the surrounding oil medium (lipid-out) and allowing time for the lipid monomers to assemble at the water–oil interface prior to positioning the microdroplets into contact.1 To this effect, droplet manipulation has been achieved using a variety of different methods including manually2 or robotically3 controlled micromanipulators, compressible PDMS substrates,4 a/c electrokinetics,5 and microfluidics.6
Fundamentally, DIBs have several advantages over the conventional methods used to assemble model membranes, such as supported lipid bilayers, black lipid membranes and liposomes. These include relative ease of assembly,1 high mechanical and temporal stability,7 the compatibility with high-throughput generation on chip and, perhaps most significantly, the ability to form interconnected 2D and 3D DIB networks.6,8–10 The assembly of DIB networks is interesting as it enables the formation of more complex systems by incorporating protein channels, pores and other cellular machinery. To this accord, DIB networks have been shown to behave as bio-batteries10 and current rectifiers11 in addition to propagating enzymatic cascades.12 There have also been advances concerning the development of DIB networks that operate in physiological (i.e. aqueous) environments, using micropipetting13,14 and microfluidic technologies.34 This is significant in the context of synthetic biology as it supports the concept that these microdroplet networks could be used to form minimal cells, micro-reactor networks, biological circuits, smart biomaterials and minimal tissues.8
To fully realise this goal, it is desirable for DIB generation strategies to have: (i) high spatial and temporal resolution for the manipulation of cell-sized microdroplets (ii) the flexibility to assemble any user-defined network either in 2D or in 3D (iii) the capability of addressing and discriminating between heterogeneous droplets to enable the assembly of complex DIB networks from multiple droplet types.
Our lab has previously shown the controlled assembly of 2D DIB networks from three alternating 40 pL droplets inside branched microfluidic channels in addition to the construction of more complex 3D DIBs networks inside a linear channel device.15 In both methods the ability to customise the network topology was dictated by the channel geometry. In another approach, Bayley and co-workers have shown the automated assembly of thousands of 65 pL droplets into complex DIB networks from two individually addressable sources by 3D printing.8 More recently, the same group has also shown the construction of 3D DIB networks using magnetic fields by inserting magnetic beads into four different manually dispensed droplet types ranging from 80–200 nL in volume.16 Although both techniques offer the ability to assemble bespoke 2D and 3D networks, a potential drawback of the 3D printing method is the inability to independently re-address individual droplets after dispensing, while the limiting factor of the magnetic bead technique is the relatively large droplet volumes. Contactless laser-based approaches to DIB formation, which exploit the fluid motion generated by either the thermocapillarity effect or Marangoni convection, have also been proposed.17,18 However, these methods offer little or no control over the positioning of individual droplets and do not allow the formation of 3D DIB networks.
Here we report a new method to assemble complex 2D and 3D DIB networks from cell-sized microdroplets <15 pL in volume (∅ < 30 μm) using a single-beam optical trap (optical tweezers). The formation of the optically assembled DIB (OptiDIB) is confirmed by demonstrating the interdroplet exchange of calcium ions through alpha haemolysin (αHL) nanopores inserted into the membrane. The novelty of our OptiDIB platform is the ability to assemble 3D DIB networks by directly trapping the microdroplets individually with the laser as opposed to exploiting the collective droplet motion triggered by laser-heating effects.17,18 While DIB formation has been achieved using the former, the lower refractive index of the aqueous droplets in relation to the oil medium prohibits direct trapping with optical tweezers and limits droplet manipulation to a ‘kicking’ effect which precludes the possibility of assembling 3D networks. Although this can be overcome by re-shaping the laser beam through either static or dynamic optical devices,19 the problem can also be circumvented entirely by adding sucrose to the droplets to reverse the refractive index contrast.20,21 Using our approach, we show that optical tweezers in combination with a motorised microscope stage can be used to drag and drop cell-sized microdroplets in 3D to assemble complex 2D and 3D OptiDIB networks with user defined connectivity for the first time. To our knowledge, the OptiDIB networks with 2D/3D architectures assembled herein are among the smallest and most complicated reported to date.
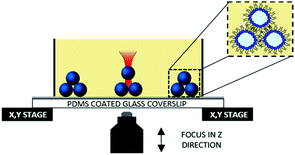
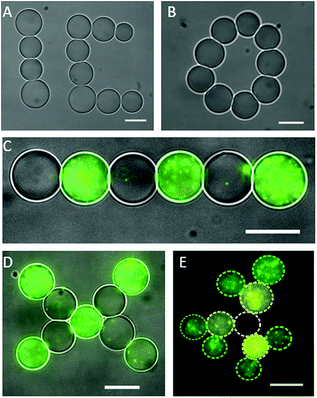
Microdroplets were generated off-chip (see ESI†) and loaded inside a well on top of a PDMS coated glass coverslip. In this configuration the well could be scanned for suitably sized microdroplets, which were individually trapped with the laser, lifted off the PDMS coated surface by displacing the objective in the Z direction and repositioned for network assembly by moving the microscope stage in the X or Y directions. Once a suitable number of microdroplets were collected, 2D or 3D OptiDIB networks were assembled in user-defined configurations by precisely positioning individual microdroplets into contact (Fig. 1). Once manipulated into contact, it was found that the droplets became attached and the interdroplet interface became deformed within a few seconds. The strength of the interdroplet bond, which we attribute to bilayer formation, was highlighted by our ability to drag the second droplet in 3D via the first, an effect that could also be reproduced for a short chain up to four microdroplets long (see ESI†). By connecting more droplets, a linear and a branched 2D OptiDIB network, “IC”, was assembled (Fig. 2A), in addition to a closed ring structure from 9 droplets (Fig. 2B). The infrequency of droplet coalescence observed during or after network assembly indicated that the OptiDIBs formed were extremely stable. It was also found that the droplets remained intact for over 24 hours provided that no droplet shrinkage was observed. Interestingly it was noted that this shrinkage phenomenon, which has been reported previously for similarly sized microdroplets in oil,22–26 was not routinely observed in our experiments. We speculate that this may be due to the inevitable variability of the water to oil ratio of the final microdroplet dispersion manually dispensed into the well.
While membrane proteins have been studied in DIB systems where both bilayer leaflets are composed of one lipid,2 or the same mixture of lipids,27 it is understood that the bilayer leaflets of almost all biological membranes are compositionally asymmetric.28 This asymmetry is thought to play a key role in cellular processes,29–31 including the gating of protein pores,32 partly via its influence on the membranes mechanical properties.35,36 Bilayer asymmetry can be achieved in DIB systems by supplying the two constituent droplets with different lipids.33 To show that asymmetric DIB networks can be assembled using our method, we prepared multiple populations of cell-sized microdroplets containing vesicles composed of different fluorescent lipids (see ESI†). The use of the appropriate filter cubes enabled the different droplets to be identified by fluorescence microscopy prior to collection for network assembly. To this effect interconnected 2D OptiDIB networks with alternating droplet types (Fig. 2C and D) and a more complex branched network from three droplet types (Fig. 2E) were constructed. The micrograph composites in Fig. 2 highlight the size/volume of the droplets (∅ < 15 μm, V < 1.8 pL), the degree of bilayer asymmetry introduced into the OptiDIB networks and the ability to assemble asymmetric OptiDIB networks in topologies which cannot be achieved using standard microfluidic techniques.
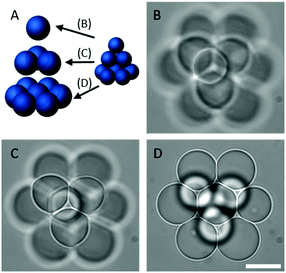
Another feature of our method which cannot be fully replicated using conventional microfluidic approaches is the ability to construct user-defined 3D DIB networks from cell-sized droplets. To illustrate this capability we assembled a 3 layer droplet pyramid by precisely positioning 11 individual droplets (∅ < 20 μm, V = <4.5 pL) into contact (Fig. 3A). While a similar structure has been reported previously, significantly larger droplets were used.16 Brightfield micrographs at each focal plane show a single microdroplet (Fig. 3B) stacked on top of a three droplet network (Fig. 3C) which sits on top of a seven droplet network (Fig. 3D). It was noted that the 3D OptiDIB networks, which were typically assembled from similarly sized microdroplets, were stable for the duration of our experiments and did not collapse. We attribute this stability to the presence of lipid bilayers at the droplet interfaces, as indicated by the deformation of the droplets in our brightfield micrographs in Fig. 3B–D.
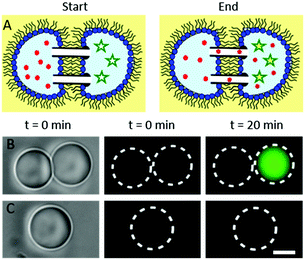
To verify the presence of lipid bilayers at the interdroplet interfaces of our OptiDIB networks, a fluorescent assay was performed using a calcium sensitive dye (Fluo-4) to detect calcium ions diffusing through αHL nanopores inserted into the membrane (Fig. 4A).13 As calcium ions cannot passively diffuse across a membrane and the assembly of αHL pores requires the presence of a lipid bilayer, the observation of a gradually increasing fluorescent signal is indicative of OptiDIB formation. To demonstrate this effect, droplets containing buffer, αHL and either (i) Fluo-4 and ethylenediaminetetraacetic acid (EDTA) or (ii) calcium chloride were prepared. Droplet pairs were subsequently assembled using the laser (Fig. 4B) and a single type (i) droplet was used as control (Fig. 4C). Fluorescent micrographs of the droplets were taken at regular intervals and analysed using ImageJ to determine both the intensity and the size (see ESI†) of the type (i) droplets. After 10 minutes the fluorescent signal of the droplet pair in Fig. 4B increased ca. 1.4× (see ESI†), while the intensity of the control droplet remained constant. A slightly larger signal was observed over the same time period for bigger droplets selected from the same preparation (see ESI†), suggesting that the signal to noise ratio is sensitive to droplet volume. Since no droplet shrinkage was observed in either instance, we attribute this increase in the fluorescent signal to the passive influx of calcium ions through αHL from the neighbouring droplet, thereby confirming DIB formation.
Given that 1.75 M sucrose has not been previously reported to block αHL pores,12 and the characteristic time for calcium ions to diffuse across a 20 μm droplet containing 2 M sucrose is ca. 8 s (see ESI†) our data suggests that the rate limiting step of the fluorescent assay is determined by the rate of ion transport across the bilayer. As the rate of ion transportation can be tuned by functionalising the bilayer with biological or synthetic nanopores, it is plausible that DIB networks could facilitate reaction cascades where both sequence and reaction rates are controlled exclusively by the properties of the DIB. This presents tremendous potential for the construction of versatile and highly regulated smart drug delivery systems, drug screening platforms, bio-sensors and minimal artificial tissues.
In conclusion, this study showcases a new technique for rapidly assembling cell-sized microdroplets into user defined 2D and 3D DIB networks using optical tweezers. Our method surpasses the requirement for a complicated optical setup, is versatile, contactless, and benefits from both high spatial and temporal resolution. This approach thus enhances the range and complexity of network geometries that can be assembled. The OptiDIB networks we report are extremely stable and, to our knowledge, are among the smallest and most intricate reported to date. These developments are especially important given that DIB platforms are increasingly being recognised as powerful architectures for fabricating micro-reactors, artificial tissues, artificial cells and bioelectric circuits and have already shown promise as the basis for a new generation of networked bio-devices.9 Given the flexibility of the technology, individual droplets can potentially be added and removed from a biological circuit, enabling for instance the addition of a power module to start or sustain a reaction and the removal of waste droplets containing the reaction products from a micro-reactor network. Ultimately, the complexity of a given network is only limited by how many types of droplets a user wishes to add to a network as the flexibility of the plug-and-play optical trapping approach enables the users to sort between them. Miniaturisation to cell-sized dimensions further increases its relevance as a platform technology in synthetic biology, and facilitates applications of OptiDIB networks for drug pharmacokinetics screens as compartmentalised delivery vehicles.
This work was supported by the EPSRC via grants EP/J017566/1, EP/K038648/1, EP/K503733/1 and by an EPSRC Doctoral Prize Fellowship awarded to YE.
References
- H. Bayley, B. Cronin, A. Heron, M. A. Holden, W. L. Hwang, R. Syeda, J. Thompson and M. Wallace, Mol. BioSyst., 2008, 4, 1191–1208 RSC.
- R. Syeda, M. A. Holden, W. L. Hwang and H. Bayley, J. Am. Chem. Soc., 2008, 130, 15543–15548 CrossRef CAS PubMed.
- A. M. El-Arabi, C. S. Salazar and J. J. Schmidt, Lab Chip, 2012, 12, 2409–2413 RSC.
- S. A. Sarles and D. J. Leo, Anal. Chem., 2010, 82, 959–966 CrossRef CAS PubMed.
- S. Aghdaei, M. E. Sandison, M. Zagnoni, N. G. Green and H. Morgan, Lab Chip, 2008, 8, 1617–1620 RSC.
- C. Stanley, K. Elvira, X. Niu, A. Gee, O. Ces and J. Edel, Chem. Commun., 2010, 46, 1620–1622 RSC.
- S. A. Sarles and D. J. Leo, Lab Chip, 2010, 10, 710–717 RSC.
- G. Villar, A. D. Graham and H. Bayley, Science, 2013, 340, 48–52 CrossRef CAS PubMed.
- M. J. Booth, V. R. Schild, A. D. Graham, S. N. Olof and H. Bayley, Sci. Adv., 2016, 2 DOI:10.1126/sciadv.1600056.
- M. A. Holden, D. Needham and H. Bayley, J. Am. Chem. Soc., 2007, 129, 8650–8655 CrossRef CAS PubMed.
- G. Maglia, A. J. Heron, W. L. Hwang, M. A. Holden, E. Mikhailova, Q. Li, S. Cheley and H. Bayley, Nat. Nanotechnol., 2009, 4, 437–440 CrossRef CAS PubMed.
- Y. Elani, R. V. Law and O. Ces, Nat. Commun., 2014, 5, 5305, DOI:10.1038/ncomms6305.
- G. Villar, A. J. Heron and H. Bayley, Nat. Nanotechnol., 2011, 6, 803–808 CrossRef CAS PubMed.
- Y. Elani, A. Gee, R. V. Law and O. Ces, Chem. Sci., 2013, 4, 3332–3338 RSC.
- Y. Elani, A. J. deMello, X. Niu and O. Ces, Lab Chip, 2012, 12, 3514–3520 RSC.
- T. Wauer, H. Gerlach, S. Mantri, J. Hill, H. Bayley and K. T. Sapra, ACS Nano, 2014, 8, 771–779 CrossRef CAS PubMed.
- S. S. Dixit, H. Kim, A. Vasilyev, A. Eid and G. W. Faris, Langmuir, 2010, 26, 6193–6200 CrossRef CAS PubMed.
- S. S. Dixit, A. Pincus, B. Guo and G. W. Faris, Langmuir, 2012, 28, 7442–7451 CrossRef CAS PubMed.
- R. M. Lorenz, J. S. Edgar, G. D. Jeffries, Y. Zhao, D. McGloin and D. T. Chiu, Anal. Chem., 2007, 79, 224–228 CrossRef CAS PubMed.
- P. M. Bendix and L. B. Oddershede, Nano Lett., 2011, 11, 5431–5437 CrossRef CAS PubMed.
- R. M. Lorenz, J. S. Edgar, G. D. M. Jeffries and D. T. Chiu, Anal. Chem., 2006, 78, 6433–6439 CrossRef CAS PubMed.
- P. B. Duncan and D. Needham, Langmuir, 2006, 22, 4190–4197 CrossRef CAS PubMed.
- F. Eslami and J. A. Elliott, J. Phys. Chem. B, 2013, 117, 2205–2214 CrossRef CAS PubMed.
- G. D. Jeffries, J. S. Kuo and D. T. Chiu, J. Phys. Chem. B, 2007, 111, 2806–2812 CrossRef CAS PubMed.
- A. Q. Shen, D. Wang and P. T. Spicer, Langmuir, 2007, 23, 12821–12826 CrossRef CAS PubMed.
- J. B. Boreyko, P. Mruetusatorn, S. A. Sarles, S. T. Retterer and C. P. Collier, J. Am. Chem. Soc., 2013, 135, 5545–5548 CrossRef CAS PubMed.
- M. S. Friddin, N. P. Smithers, M. Beaugrand, I. Marcotte, P. T. Williamson, H. Morgan and M. R. de Planque, Analyst, 2013, 138, 7294–7298 RSC.
- G. van Meer, D. R. Voelker and G. W. Feigenson, Nat. Rev. Mol. Cell Biol., 2008, 9, 112–124 CrossRef CAS PubMed.
- G. S. Attard, R. H. Templer, W. S. Smith, A. N. Hunt and S. Jackowski, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 9032–9036 CrossRef CAS PubMed.
- T. Pomorski, R. Lombardi, H. Riezman, P. F. Devaux, G. van Meer and J. C. Holthuis, Mol. Biol. Cell, 2003, 14, 1240–1254 CrossRef CAS PubMed.
- B. Fadeel and D. Xue, Crit. Rev. Biochem. Mol. Biol., 2009, 44, 264–277 CrossRef CAS PubMed.
- E. Perozo, A. Kloda, D. M. Cortes and B. Martinac, Nat. Struct. Mol. Biol., 2002, 9, 696–703 CAS.
- W. L. Hwang, M. Chen, B. Cronin, M. A. Holden and H. Bayley, J. Am. Chem. Soc., 2008, 130, 5878–5879 CrossRef CAS PubMed.
- Y. Elani, C. I. Xavier, J. B. Edel, R. V. Law and O. Ces, Chem. Commun., 2016, 52, 5961–5964 RSC.
- Y. Elani, S. Purushothaman, P. J. Booth, J. M. Seddon, N. J. Brooks, R. V. Law and O. Ces, Chem. Commun., 2015, 51, 6976–6979 RSC.
- L. Lu, W. J. Doak, J. W. Schertzer and P. R. Chiarot, Soft Matter, 2016 10.1039/C6SM01349.
Footnote |
| † Electronic supplementary information (ESI) available: Details of lipid preparation, optical setup, bilayer assembly and fluorescent assay in addition to videos of DIB and DIB network formation. See DOI: 10.1039/c6sm01357k |
| This journal is © The Royal Society of Chemistry 2016 |