Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Structures of malonic acid diamide/phospholipid composites and their lipoplexes†‡
Christopher
Janich
*ab,
Stephanie
Taßler
c,
Annette
Meister
d,
Gerd
Hause
e,
Jens
Schäfer
b,
Udo
Bakowsky
b,
Gerald
Brezesinski
c and
Christian
Wölk
*a
aMartin Luther University Halle-Wittenberg, Institute of Pharmacy, Wolfgang-Langenbeck-Strasse 4, 06120 Halle, Saale, Germany. E-mail: christopher.janich@pharmazie.uni-halle.de; christian.woelk@pharmazie.uni-halle.de; Fax: +49-345-55-27018; Fax: +49-345-55-27018; Tel: +49-345-55-25077 Tel: +49-345-55-25078
bPhilipps University Marburg, Department of Pharmaceutical Technology and Biopharmacy, Ketzerbach 63, 35037 Marburg, Germany
cMax Planck Institute of Colloids and Interfaces, Science Park Potsdam-Golm, Am Mühlenberg 1, 14476 Potsdam, Germany
dMartin Luther University Halle-Wittenberg, Institute of Chemistry, Physical Chemistry and Institute of Biochemistry and Biotechnology, von-Danckelmann-Platz 4, 06120 Halle, Saale, Germany
eMartin Luther University Halle-Wittenberg, Biocenter, Weinbergweg 22, 06120 Halle, Saale, Germany
First published on 30th May 2016
Abstract
As a continuation of previous work, the self-assembly process of cationic lipid formulations in the presence and absence of DNA was investigated with respect to binary lipid mixtures suitable as polynucleotide carrier systems. The lipid blends consist of one malonic-acid-based cationic lipid with a varying alkyl chain pattern, either N-{6-amino-1-[N-(9Z)-octadec-9-enylamino]-1-oxohexan-(2S)-2-yl}-N′-{2-[N,N-bis(2-aminoethyl)amino]ethyl}-2-hexadecylpropandiamide (OH4) or N-[6-amino-1-oxo-1-(N-tetradecylamino)hexan-(2S)-2-yl]-N′-{2-[N,N-bis(2-aminoethyl)amino]ethyl}-2-hexadecylpropandiamide (TH4), and one neutral co-lipid, either 1,2-di-[(9Z)-octadec-9-enoyl]-sn-glycero-3-phosphocholine (DOPE) or 1,2-di-(hexadecanoyl)-sn-glycero-3-phosphocholine (DPPC). Although the cationic lipids exhibit only slight differences in their structure, the DNA transfer efficiency varies drastically. Therefore, self-assembly was studied in 3D systems by small- and wide-angle X-ray scattering (SAXS and WAXS) and transmission electron microscopy (TEM) as well as in 2D systems by infrared reflection–absorption spectroscopy (IRRAS) on Langmuir films. The investigated lipid mixtures show quite different self-assembly in the absence of DNA, with varying structures from vesicles (OH4/DOPE; TH4/DOPE) and tubes (TH4/DOPE) to discoid structures (OH4/DPPC; TH4/DPPC). Twisted ribbons and sheets, which were stabilized due to hydrogen-bond networks, were found in all investigated lipid mixtures in the absence of DNA. The addition of DNA leads to the formation of lamellar lipoplexes for all the investigated lipid compositions. The lipoplexes differ in crucial parameters, such as the lamellar repeat distance and the spacing between the DNA strands, indicating differences in the binding strength between DNA and the lipid composition. The formation of associates with an ideal charge density might emerge as a key parameter for efficient DNA transfer. Furthermore, the structures observed for the different lipid compositions in the absence of DNA prepare the way for other applications besides gene therapy.
1 Introduction
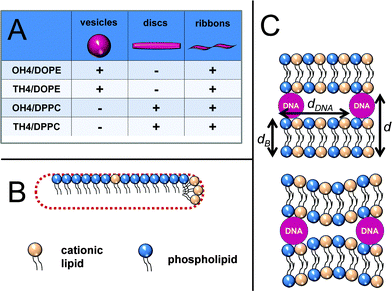
Since the beginning of the gene therapy treatment concept in medicine, efficient approaches in transferring nucleic acids (e.g. siRNA, mRNA, plasmids, or antisense oligonucleotides) into cells were developed.1,2 For this purpose, different polynucleotide delivery systems (vectors) are required, which are divided into two major classes: viral and non-viral vectors.3,4 The most widely used non-viral vectors are lipid-based delivery systems.5 Lipid-mediated polynucleotide transfer is referred to as lipofection, which was initially described by Felgner et al.6 Lipid-based vectors are characterized by facile and variable preparation, a low immunogenic potential, and a high loading capacity.7 Moreover, the utilization of biodegradable lipids enables the preparation of vectors of relatively low toxicity.8 Nevertheless, there are still some disadvantages, such as a lower transfection efficiency compared to viral vectors. To develop new non-viral gene delivery devices with a high gene transfer efficiency combined with low toxic effects, fundamental knowledge is needed about the structure/activity correlations of the lipid DNA complexes (lipoplexes), and their ability to interact with cell membranes, to be internalized in an efficient manner and to release nucleic acids from the lipoplex.9–13 Furthermore, the determination of the physical characteristics of the lipoplex, as well as the lipid composition itself, is indispensable.14 Therefore numerous investigations with multidisciplinary approaches are necessary.A promising class of cationic lipids are the malonic acid diamides of the second generation.15 Recently, we published physical–chemical parameters of composites of cationic malonic acid diamides (cytofectine) and phospholipids (co-lipid), and of their lipoplexes, and reported on the influence on the DNA transfer efficiency.16,17 For these structure-function studies, the two malonic acid diamides, N-{6-amino-1-[N-(9Z)-octadec-9-enylamino]-1-oxohexan-(2S)-2-yl}-N′-{2-[N,N-bis(2-aminoethyl)amino]ethyl}-2-hexadecylpropandiamide (OH4) and N-[6-amino-1-oxo-1-(N-tetradecylamino)hexan-(2S)-2-yl]-N′-{2-[N,N-bis(2-aminoethyl)amino]ethyl}-2-hexadecylpropandiamide (TH4), have been used. The cationic lipids exhibit identical head groups but differ in the alkyl chain pattern (Fig. 1). Binary mixtures of these cationic lipids with the phospholipids, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) or 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), in the molar ratio 1/1 exhibit quite different transfection efficiencies: OH4/DOPE was the most effective one, while TH4/DOPE, OH4/DPPC and TH4/DPPC showed only minor transfection efficiencies. The key question arose: why does the OH4/DOPE mixture exhibit such high transfection efficiency in comparison to the other investigated lipid mixtures? Previous research demonstrated that the length, fluidity and saturation degree of the aliphatic chain of the cationic lipid and the type of phospholipid play a major role in the lipid mixing and influence the charge density.16 Moreover, it was demonstrated that the optimal binding strength between the lipid composites and DNA plays an important role in efficient gene transfer into cells, whereby premature DNA release by biological agents is a major parameter influencing the transfection efficiency.17
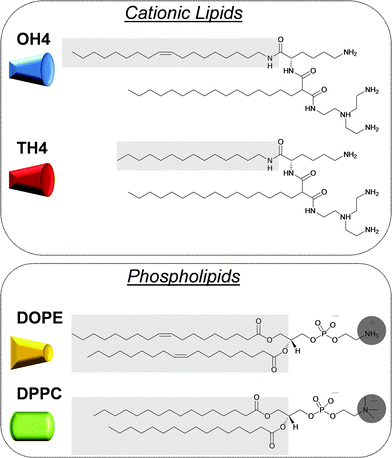 | ||
| Fig. 1 Structures of the cationic lipids and phospholipids used in this study. Structural differences are highlighted using a grey background. The cationic lipids favour a positive curvature. DOPE has negative curvature effects, whereas DPPC forms planar assemblies. The determination of the packing parameter and the resulting preferred curvature was described earlier.16 | ||
The aim of this paper is the investigation of structures formed by the four lipid mixtures and their complexes with DNA. For this purpose, small- and wide-angle X-ray scattering (SAXS and WAXS) experiments were performed. Additional investigations by transmission electron microscopy (TEM) allowed the determination of structures resulting from self-assembling of the amphiphiles in the presence and absence of DNA. Furthermore, infrared reflection–absorption spectroscopy (IRRAS) experiments were performed to investigate Langmuir monolayer models.18 The investigations show that the exchange of an oleyl chain by a tetradecyl chain in the cationic lipids (OH4→TH4) drastically influences the self-assembly behaviour of the lipid formulations, resulting in a decrease in the DNA transfer efficiency. Furthermore, the exchange of the co-lipid DOPE by DPPC results in a quite different self-assembly behaviour of the investigated binary lipid mixtures. The self-assembling in the presence of DNA results in lamellar lipoplexes for all four investigated lipid composites. Nevertheless, the structural parameters of the lipoplexes, which are also a result of the binding strength between the lipid mixtures and the DNA, are clearly different.
2 Experimental
2.1 Materials
Unless otherwise stated, all materials were purchased from Sigma-Aldrich. The buffers were prepared with Milli-Q Millipore water with a specific resistance of 18.2 MΩ cm. The 2-(N-morpholino)ethanesulfonic acid (MES) buffer was adjusted to pH 6.5 and filtered through a 0.2 mm cellulose acetate membrane before use. HBr buffer was prepared from HBr acid and adjusted to pH 3 using 1,4-diazabicyclo-(2,2,2)-octane. Lyophilised calf thymus (ct) DNA and ctDNA solution (1 mg mL−1) was purchased from Sigma-Aldrich. The synthesis of TH4 (N-[6-amino-1-oxo-1-(N-tetradecylamino)hexan-(2S)-2-yl]-N′-{2-[N,N-bis(2-aminoethyl)amino]ethyl}-2-hexadecylpropandiamide) and OH4 (N-{6-amino-1-[N-(9Z)-octadec-9-enylamino]-1-oxohexan-(2S)-2-yl}-N′-{2-[N,N-bis(2-aminoethyl)amino]ethyl}-2-hexadecylpropandiamide) was described previously.15,19 DOPE and DPPC were purchased from Avanti Polar Lipids (Alabaster, AL, USA).2.2 SAXS/WAXS
Lipids were dissolved in chloroform/methanol (8/2, v/v) and, for the binary mixtures, were combined at a molar ratio of 1/1. The solutions were transferred to 0.1 mL micro-insert vials (VWR™ International) and the solvent was evaporated under a continuous flow of argon, yielding a film. The film was dried for 24 h under vacuum. Lipid samples were prepared as 10 wt% lipid dispersions in buffer (0.1 M MES, pH 6.5 or HBr buffer, pH 3). To ensure complete hydration, the dispersions were vortexed and heated up to 55 °C (above the phase transition) twice. Lipid/DNA-complexes at an N/P (number of NH2 functions of the cationic lipid/number of PO4 functions in the DNA) ratio of 4 were prepared by adding ctDNA stock solution in MES buffer (0.1 M, pH 6.5). For all SAXS measurements, the dispersions were transferred to glass capillaries (diameter 1.5 mm, borosilicate glass, WJM Berlin, Germany) and stored for at least 3 days at 4 °C before measurement.The High Brilliance Beamline ID02 (ESRF, France) was used for the measurements in HBr buffer at pH 3. The energy of the incident X-ray beam was 12.5 keV (λ = 0.992 Å), the beam size was about 100 μm, and the sample-to-detector distance was 1.2 m. The SAXS patterns were collected with a 4 FT-CCD detector (Rayonix MX-170HS). For SAXS, a q range from 0.006 Å−1 to 0.65 Å−1 with a detector resolution of 3 × 10−4 Å−1 was used. WAXS data were obtained in a q range from 0.72 Å−1 to 5.1 Å−1. To avoid radiation damage, each sample was measured with 10 frames with an exposure time of 0.05 s per frame. For data analysis, the average of all 10 frames was used. The angular calibration of the SAXS detectors was performed using silver behenate powder as reference, and for the WAXS detector (Rayonix LX-170HS), p-bromobenzoic acid was used. The temperature was adjusted with a Huber Unistat thermostatic bath with an accuracy of ±0.1 °C. Experiments have been performed at 25 °C. The sealed glass capillaries containing the lipid dispersions were well-positioned in a Peltier controlled automatic sample changer. The real-space repeat distance, d, of the lattice planes was calculated from the position of the first diffraction peak. The collected 2D powder diffraction spectra were treated with BH plots (Macro in MathLab) and analysed using Origin 8.6.
The μSPOT beamline (BESSY II, Germany) was used for the measurements in MES buffer at pH 6.5. The energy of the incident X-ray beam was 12.0 keV (λ = 1.033 Å), the beam size was defined by a pinhole with a diameter of 100 μm, followed by a guard pinhole with a diameter of 150 μm, and the sample-to-detector distance was 54.1 cm. The SAXS and WAXS patterns were collected with a MarMosaic 225 detector (MarUSA, Evanston, USA), consisting of 9 independent 16-bit CCD chips connected to a phosphor screen of 225 mm edge length (pixel size 73.24 μm) with fiber optic tapers. Experiments have been performed at 25 °C (Peltier element). The angular calibration of the SAXS detectors was performed using silver behenate powder as reference, and for WAXS, p-bromobenzoic acid was used. The collected 2D powder diffraction spectra were treated with FIT2D V16.041 and analysed in Origin 8.6.
The diffraction peaks were fitted with the Voigt-function. The GAP fits for the lipid mixtures and pure cationic lipid dispersions were performed with GAP 1.3, written by Georg Pabst,20–22 using the no-structure factor mode (no Bragg peaks, but pure diffuse scattering from positionally uncorrelated bilayers; detailed information about the GAP fit is given in the ESI‡).
2.3 IRRAS
Infrared reflection–absorption spectra were recorded on a Vertex 70 FTIR spectrometer (Bruker, Ettlingen, Germany). The setup included a film balance (R&K, Potsdam, Germany) and an external air–water reflection unit, XA-511 (Bruker). A sample trough with two movable barriers, for compressing the lipid monolayer, and a reference trough (bare subphase) allowed the fast recording of sample and reference spectra by a shuttle technique. The infrared beam was focused on the liquid surface by a set of mirrors. The angle of incidence normal to the surface can be varied by means of moveable arms in the range between 30° and 72°. A KRS-5 wire grid polarizer was used to polarize the IR radiation, either in a parallel (p) or perpendicular (s) direction. After reflection from the surface, the beam was directed to a narrow-band mercury cadmium telluride detector (MCT) cooled with liquid nitrogen. Reflectance–absorbance spectra were obtained by using −log(R/R0), where R is the reflectance of the film-covered surface and R0 is the reflectance of the same subphase without the film. For each single-beam spectrum, 200 scans (s-polarized light) or 400 scans (p-polarized light) were added with a scanning velocity of 20 kHz and a resolution of 8 cm−1, apodized using the Blackman–Harris three-term function, and fast Fourier transformed after one level of zero filling.23 For data analysis, spectra obtained with s- and p-polarized light and an angle of incidence of 40° were used. All spectra were corrected for atmospheric interference using the OPUS software and baseline corrected using the spectral subtraction software. The spectra were not smoothed. To determine the exact position of the bands, a Lorentzian curve was fitted to the data points.2.4 CryoTEM
Lipid dispersions were prepared using the film hydration procedure. Accordingly, the lipids were dissolved in chloroform/methanol (9/1, v/v) and combined at the appropriate molar ratio for the mixtures. Lipid films were obtained by evaporating the solvent for 1 h at 200 mbar and for a further 3 h at 10 mbar. After evaporation, 10 mM MES buffer at pH 6.5 was added to give a stock dispersion (2 mg mL−1). The lipid formulations were incubated at 50 °C while shaking (1400 rpm, 30 min), followed by sonication (37 kHz, 50 °C, 3 min). Lipoplexes were prepared in a one-step mixing procedure by adding ctDNA stock solution to the lipid formulation. The samples were incubated for 15 min at 25 °C. Vitrified specimens were prepared using a blotting procedure, performed in a chamber with controlled temperature and humidity using an EM GP grid plunger (Leica, Wetzlar, Germany). The sample solution (6 μL) was placed onto an EM grid coated with a holey carbon film (Cflat, Protochips Inc., Raleigh, NC). Excess solution was then removed by blotting with a filter paper to leave a thin film of the solution spanning the holes of the carbon film on the EM grid. Vitrification of the thin film was achieved by rapid plunging of the grid into liquid ethane held just above its freezing point. The vitrified specimen was kept below 108 K during storage, transferred to the microscope and investigated. Specimens were examined with a Libra 120 Plus transmission electron microscope (Carl Zeiss Microscopy GmbH, Oberkochen, Germany), operating at 120 kV. The microscope was equipped with a Gatan 626 cryotransfer system. Images were acquired using a BM-2k-120 dual-speed on-axis SSCCD camera (TRS).2.5 Negative stain TEM
Lipoplex dispersions were prepared as described above for the cryoTEM experiments and diluted with MES buffer to a concentration of 0.05 mg mL−1. The negatively stained samples were prepared by spreading the dispersion (5 μL) onto a Cu grid coated with a formvar film (Plano, Wetzlar, Germany). After 1 min, excess liquid was removed by blotting with filter paper and 1% aq. uranyl acetate (5 μL) was placed onto the grid and drained off after 1 min. The dried specimens were examined using an EM 900 transmission electron microscope (Carl Zeiss Microscopy GmbH, Oberkochen, Germany). Micrographs were acquired using an SSCCD SM-1k-120 camera (TRS, Moorenweis, Germany).3 Results and discussion
3.1 Characterization of the aggregation behaviour of pure cationic lipids
While the chemical structures of OH4 and TH4 vary in only one alkyl chain, by replacing an oleyl chain with a tetradecyl chain (Fig. 1), the aggregation behaviour in aqueous dispersions is quite different. OH4 assembles into a 3D sponge-like network (Fig. 2A). The dominant aggregates of TH4 in the aqueous dispersion are sheet-like layers, clearly visible in cryoTEM by the wrinkles that result from sample preparation (Fig. 2B, white arrows).The SAXS patterns of OH4 at pH 3 and pH 6.5 show diffuse scattering curves resulting from weakly correlated bilayers, which are also comparable with spectra described for the L3 sponge phase.24 The reflexes at pH 3 are more sharp and three reflexes are detected (Fig. 2C). The reflex at q = 1.37 nm−1 (L001) results in d = 45.9 Å. Additional reflexes appear at q = 4.05 nm−1 and q = 4.42 nm−1. Comparable reflexes can also be observed at pH 6.5, but are less clear due to the lower signal-to-noise ratio (Fig. 2E). The treatment of the SAXS data at pH 6.5 with the GAP fit (Fig. 2G and H) yields a bilayer thickness (dB) of (47.5 ± 6.9) Å. To obtain information about the alkyl chain state, WAXS experiments were performed. The WAXS pattern at pH 3 shows 3 peaks (Fig. 2D). The peak at q11 = 14.84 nm−1, taken as an indication of hexagonal chain packing, leads to a cross-sectional area of A = 2/(31/2shk2) = 20.7 Å2. The peaks at qH-bond-1 = 12.78 nm−1 and qH-bond-2 = 13.29 nm−1 can be attributed to a periodic H-bond lattice with d = 4.92 Å (H-bond-1) and d = 4.73 Å (H-bond-2). The distances fit with the distances of N–H⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C hydrogen bonds of β-sheets of proteins,19,25,26 and were described earlier in the literature for lipids with a related backbone.27 A comparable WAXS pattern can be observed at pH 6.3, but is less clear due to the lower signal-to-noise ratio (Fig. 2F): one peak at q11 = 15.25 nm−1, resulting in A = 19.6 Å2 and a second weak peak at qH-bond = 13.04 nm−1 with d = 4.82 Å. DSC experiments with OH4 in MES buffer at pH 6.5 show no phase transition between 5 °C and 80 °C.16 Based on the presented WAXS data, the lipid is in the gel state within this temperature range, contrary to the assumption made in previous research.16
C hydrogen bonds of β-sheets of proteins,19,25,26 and were described earlier in the literature for lipids with a related backbone.27 A comparable WAXS pattern can be observed at pH 6.3, but is less clear due to the lower signal-to-noise ratio (Fig. 2F): one peak at q11 = 15.25 nm−1, resulting in A = 19.6 Å2 and a second weak peak at qH-bond = 13.04 nm−1 with d = 4.82 Å. DSC experiments with OH4 in MES buffer at pH 6.5 show no phase transition between 5 °C and 80 °C.16 Based on the presented WAXS data, the lipid is in the gel state within this temperature range, contrary to the assumption made in previous research.16
TH4 shows scattering patterns similar to OH4 in SAXS and WAXS, but with slight differences. A very broad halo (L001) was observed in SAXS at pH 3 and 6.5 (Fig. 2C and E). The GAP fit of the SAXS curve at pH 6.5 results in dB = (47.7 ± 2.3) Å (Fig. 2I and J). Additionally, a distinct reflex is found at q = 4.24 nm−1 at pH 3 in concordance with the OH4 SAXS pattern. The reflex is also observable at pH 6.5. The WAXS pattern of TH4 shows more reflexes in comparison with OH4. At pH 3, the reflex of the chain lattice with q11 = 15.25 nm−1 indicates a hexagonal chain lattice with A = 19.6 Å2, assuming that the reflex at q = 16.52 nm−1 (d = 3.8 Å) results, not from an orthorhombic chain lattice, but rather from the hydrogen-bond-stabilized spacing between two lipid backbones. This thesis is supported by 3 facts: (i) the CH2 deformation (δ CH2) band found in the IRRAS experiments (see section below) appears as a single peak (see Fig. 3D) and is not split (typical for orthorhombic chain lattice).28 (ii) The calculated repeat distance of d = 3.80 Å fits with the Cα–Cα-spacing of β-sheet structures in peptides.29 (iii) The peaks attributed to a periodic H-bond lattice, as described for OH4, are also observable: qH-bond-1 = 12.91 nm−1 (d = 4.87 Å) and qH-bond-2 = 13.43 nm−1 (d = 4.68 Å). Nevertheless, due to the absence of the peak attributed to Cα–Cα in the WAXS pattern of OH4, while the signals qH-bond-1 and qH-bond-2 occur in the patterns of both lipids, and due to the discrepancies found between WAXS and IRRAS (see section below), an orthorhombic chain lattice with q11 = 15.25 nm−1 and q02 = 16.52 nm−1 cannot be excluded. This would result in a cross-sectional area of A = 18.6 Å2 and a unit cell perpendicular to the chain axis of 4.9–7.6 Å (such unit cell values were also described for a lipid with a related backbone27 and are close to the tight herringbone arrangement of hydrocarbon chains (5.0–7.5 Å)).30 At pH 6.5, three reflexes are detected in WAXS: one peak at qH-bond = 12.98 nm−1 (reflex of repeating distances caused by hydrogen bonds), one at q11 = 15.35 nm−1, and one at q = 16.50 nm−1, comparable to the scattering curve at pH 3.
In order to obtain additional information about possible hydrogen-bond formation and the phase state of the alkyl chains in 2D models, IRRAS experiments were performed on Langmuir monolayers of OH4 and TH4 (Fig. 3). The amide groups of the lipids are H-bond acceptors and donors, besides the protonated amino groups, which act as H-bond donors. Therefore, the position of the amide I and amide II bands gives evidence for H-bond formation. H-Bonds result in a bathochromic shift of the amide I band (in the general carbonyl stretching vibration) and a hypsochromic shift of the amide II band (general NH bending vibration).31,32 In the range between 10 and 34 mN m−1, a weak amide I band was detected from 1646 cm−1 to 1657 cm−1 for monolayers of OH4 (Fig. 3A and Fig. S1, ESI‡) and from 1648 cm−1 to 1650 cm−1 for TH4 (Fig. 3D and Fig. S2, ESI‡), supporting the existence of a hydrogen-bond network for both lipids.28,33 The amide II band shows high intensity and is split into two bands [Fig. 3A and D, described as amide II (1) and (2)]. For OH4 monolayers, the peak positions of the amide II band range from 1543 cm−1 to 1537 cm−1 [amide II (1)] and from 1531 cm−1 to 1527 cm−1 [amide II (2)] (see Fig. 3A and Fig. S1, ESI‡), and for TH4 from 1542 cm−1 to 1532 cm−1 [amide II (1)] or from 1524 cm−1 to 1519 cm−1 [amide II (2)] (see Fig. 3D and Fig. S2, ESI‡). The values of the amide II (1) bands also indicate the formation of hydrogen bonds.31–34 The position of the second peak of the amide II band [amide II (2)] indicates that the N–H groups partly participate in hydrogen bonds because free N–H groups would result in a maximum at 1510 cm−1.32,34 The IRRAS experiments with p-polarized light yield comparable band positions (Fig. S3, ESI‡). Additional evidence for the NH-groups of the amide backbone being involved in hydrogen bonds is given by the position of the weak amide A band (Fig. S4, ESI‡).23,28 Nevertheless, the formation of intermolecular H-bonds between the lipids cannot be confirmed because hydrogen bonds between N–H functions and water will also be detected. However, the above presented WAXS data gives strong evidence for the thesis of a hydrogen-bond lattice between the lipid head groups.
The IRRAS signals in the CH stretching region (Fig. 3B and E) permit conclusions about the organization of the hydrocarbon chains, providing a complement to the WAXS data in the 3D model. The data show that upon increasing the lateral surface pressure from 10 mN m−1 to 34 mN m−1, the band positions of the symmetric and asymmetric methylene stretching vibrations shift to lower wavenumbers. The bathochromic shift occurs steadily with increasing pressure. The values at 10 mN m−1 are typical for chains in the liquid expanded (LE, high amount of gauche conformation in the chain) phase state for both lipids. The shift to lower wavenumbers during compression indicates an increase in trans conformations in the alkyl chains. Taking into account the fact that the pressure-area isotherms of the monolayer show no phase transition,16 the lipids are still in the LE phase. This fits with the wavenumbers of both CH2 stretching vibrations, which are too high for lipid monolayers in the liquid condensed (LC) phase (typical values of the LC phase are around 2920 cm−1 to 2917 cm−1)35–37 in the case of both lipids. The experiments with p-polarized light yield comparable results (Fig. S5, ESI‡). The CH2 deformation band appears as a single peak at all investigated pressures for both lipids, indicating oblique or hexagonal chain packing (Fig. 3A and D). Obviously, there is a discrepancy between IRRAS (fluid chains in the monolayer model) and the WAXS data (distinct signals of ordered alkyl chains in the gel state). One explanation could be a different arrangement of alkyl chains in the lipid monolayers compared to the lipid bilayers. For instance, a (partial) interdigitation of the alkyl chains in the lipid bilayers is possible, considering the larger distance between the alkyl chains, compared to the sn-1,2-phospholipids. Such interdigitation is described for 1,3-alkylated phospholipids, which also exhibit a larger distance between the alkyl chains.38,39 Zumbuehl and co-workers also described a lipid (1,3-dipalmitamidopropan-2-phosphocholine) that shows an LE/LC phase transition above 35 mN m−1 at 25 °C as a Langmuir monolayer and an interdigitated lamellar gel phase in the bulk (3D system).31,39 Also, the small bilayer thickness of around 47 Å for both lipids indicates an interdigitated gel phase.
DOPE adopts the HII phase at room temperature because of the small head group. The gel to Lα transition is at around −8 °C, while the Lα to HII transition occurs at around 10 °C.40,41 Thus, the molecular shape induces negative curvature effects. DPPC favours a lamellar ordering because of its cylindrical molecular shape (Lβ′ to Pβ′ transition at 34.2 °C and Pβ′ to Lα transition at 41.7 °C).42,43 We could also prove the existence of the HII phase for DOPE and the Lβ′ phase for DPPC in MES buffer at pH 6.5 and 25 °C (Fig. S6, ESI‡).
3.2 Characterization of cationic lipid/DOPE mixtures in the presence and absence of DNA
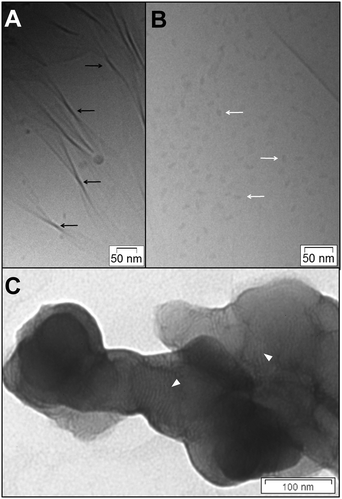
DOPE was used as co-lipid in the cationic lipid formulations because of its often discussed transfection efficiency enhancing effect.44 The possibility of DOPE-containing lipoplexes adopting inverse hexagonal mesophases is currently discussed as a reason for the enhancing effect.45,46 Aqueous dispersions of the binary mixture OH4/DOPE 1/1 (n/n) were investigated by cryoTEM. Three types of aggregates are observed: unilamellar vesicles (Fig. 4A and B, black arrows), ribbons (Fig. 4B and C, white arrows), and sheets (Fig. S7, ESI‡ black triangles). The determined diameters of the vesicles are in the range 200 nm (not shown; the biggest diameter in Fig. 4A and B is 100 nm) to 30 nm (liposomes with high curvature). The diameter determined from the cryoTEM images fits perfectly with the main particle size population determined by DLS, which was reported recently.16 The ribbons are clearly visible because of the twists. Such structures were described in the literature earlier as t-ribbons (twisted ribbons).47 The sheets are clearly visible due to the wrinkles. The existence of three different aggregates indicates phase-separation processes (a phase building curved bilayers that result in vesicles and a phase building planar aggregates such as ribbons and sheets), that could not be demonstrated for these lipid mixtures until now.16,17 The addition of DNA changes the structure of the lipid aggregates, resulting in lipoplexes with lamellar substructures (parallel ordering) (Fig. 4D, white triangles). The measured distance between the repetitive units of the lamellae is around 5 nm. No other structures besides the lipoplexes could be detected.The SAXS pattern of aqueous OH4/DOPE 1/1 (n/n) dispersions confirms the structural changes after addition of DNA. The OH4/DOPE 1/1 (n/n) mixture without DNA results in a diffuse scattering curve, an indication of weakly correlated bilayers (Fig. 5A). The weak correlation results from the repulsive forces between the positively charged lipid bilayers and is in line with the cryoTEM images, showing no multilamellar lipid aggregates without DNA (Fig. 4A–C). The GAP fit results in a bilayer thickness of (55.6 ± 1.5) Å (Fig. 5I and J). DNA has an ordering effect on the lipid mixture. Distinct reflexes appear, which are typical for the well-described Lcα lipoplex structure.48 A one-dimensional DNA lattice is sandwiched between lipid bilayers, resulting in three reflexes: q001 = 0.91 nm−1, q002 = 1.82 nm−1, and qDNA = 1.36 nm−1. This results in a lamellar repeat distance of dL = 69.0 Å (lipid bilayer with additional layer of bound DNA) and a DNA distance of dDNA = 46.2 Å (distance between the parallel ordered DNA strands of the one-dimensional DNA lattice). The WAXS patterns in the presence and absence of DNA are comparable (Fig. 5B). A weak reflex at q = 15.39 nm−1 was detected. This reflex is comparable with the qhex reflex that was found for pure OH4 (Fig. 2F). Because DOPE shows no distinct reflex in WAXS (only a halo typical for fluid alkyl chains), the reflex can result from both lipids in the mixture or, which is more likely, phase-separated OH4.
The TH4/DOPE 1/1 (n/n) mixture forms various aggregates in an aqueous dispersion at pH 6.5, which were visualised by cryoTEM images (Fig. 6A–C). Lipid tubes and vesicles are the dominating structures. Branched tubes (Fig. 6A, white triangles), elongated tubes without branching (Fig. 6B, black triangle), constricted tubes (Fig. 6B, grey triangles), vesicular structures with tubes (Fig. 6B, white triangles), small unilamellar vesicles (SUV) with diameters between 30 nm and 100 nm (Fig. 6A and D, black arrows), and deformed large unilamellar vesicles (LUV) (Fig. 6C, white arrow), with diameters between 400 nm and 1 μm, are present. The cross section of the tubes is between 40 nm and 110 nm. Exceptions are tubes with a cross section between 200 nm and 400 nm, for instance in Fig. 6A with the different embedded structures. Ribbons [slim ribbons with a width of 30 nm to 50 nm (Fig. 6A and D, black triangles), broad ribbons with a width around 150 nm (Fig. 6A, grey triangle)] and sheets (Fig. 6C, grey triangles) are also found as aggregates. Subsequently, demixing occurs, effecting a phase of curved bilayers and a phase of planar systems comparable to the OH4/DOPE 1/1 (n/n) mixture, with the difference being that the curved bilayers tend to have varying curvatures. Possible explanations will be discussed later. The lipoplex structures were investigated by TEM and are presented in Fig. 6E (cryoTEM could not be performed due to the instability of the sample in the electron beam). The aggregates show a lamellar ordered substructure comparable to OH4/DOPE 1/1 (n/n) lipoplexes (Fig. 6E, white arrow). The lamellar repeat distance is between 6 nm and 7 nm.
The SAXS pattern of aqueous TH4/DOPE 1/1 (n/n) dispersions (Fig. 5C) shows a diffuse scattering curve. The weak correlation fits with the cryoTEM images, showing no multilamellar aggregates (Fig. 6A–D). The GAP fit results in a bilayer thickness of (55.2 ± 1.6) Å (Fig. 5K and L). After addition of DNA, yielding an N/P-ratio of 4, several Bragg peaks appear (Fig. 5C). The reflexes of the layer ordering of the Lcα phase are present until the 6th order (q001 = 0.90 nm−1, q002 = 1.80 nm−1, q003 = 2.70 nm−1, q004 = 3.60 nm−1, q005 = 4.50 nm−1, and q006 = 5.40 nm−1), resulting in a lamellar repeat distance of d = 69.8 Å. Furthermore, the reflex resulting from the DNA lattice exists with qDNA = 1.59 nm−1, yielding a DNA rod distance of dDNA = 39.5 Å. This indicates tighter DNA packing, as observed for OH4/DOPE 1/1 (n/n) lipoplexes. The existence of the Lcα phase of TH4/DOPE 1/1 (n/n) lipoplexes is in line with the lamellar substructure found for lipoplexes in the TEM experiments (Fig. 6E). Two additional peaks appear, which indicate a second lamellar phase: q001 = 0.66 nm−1, and q002 = 1.32 nm−1. The calculated repeat distance is very large, with d = 95.2 Å. The pattern may be a result of lamellar lipid aggregates without DNA, with a huge water layer because of the repulsing forces of the cationic bilayers. But also, the existence of a cubic phase cannot be excluded. The WAXS data for the TH4/DOPE 1/1 (n/n) dispersion show two reflexes at q = 15.39 nm−1 and q = 16.63 nm−1 (Fig. 5D). TH4/DOPE 1/1 (n/n) lipoplexes with N/P = 4 show a comparable diffraction pattern with higher intensity (q = 15.39 nm−1 and q = 16.63 nm−1) and an additional reflex at qH-bond = 13.05 nm−1, resulting from a hydrogen-bond lattice, as described above for TH4 dispersions. It is impossible to allocate the reflexes to a distinct phase, but the WAXS pattern is comparable to the pattern found for TH4 alone. Therefore, it is most likely that the reflexes result from phase-separated TH4. An ideal mixture with DOPE would break the hydrogen-bond network of the TH4 molecules.
3.3 Characterization of cationic lipid/DPPC mixtures in the presence and absence of DNA
The OH4/DPPC 1/1 (n/n) mixture forms two different types of aggregates: discoid structures and twisted ribbons (Fig. 7A and B; Fig. S8, ESI‡). The stacks of discs were also seen in AFM images (Fig. S14, ESI‡). The addition of DNA to the lipid mixture yields lipoplexes with a lamellar substructure coexisting with the ribbons (Fig. 7C) and the lipid discs (Fig. 7D).Fig. 5E shows the SAXS pattern of hydrated OH4/DPPC 1/1 (n/n) samples at pH 6.5 and 25 °C in the presence and absence of DNA. OH4/DPPC 1/1 (n/n) dispersions without DNA show a broad Bragg peak (uncorrelated bilayers, fits with the TEM investigations). In contrast to the SAXS patterns of cationic lipid/DOPE mixtures without DNA (Fig. 5A and C), the broad reflex shows higher intensity and is accompanied by a more distinct undulation at higher q values in the SAXS pattern (Fig. 5E). An explanation is that the uncorrelated aggregates are membrane discs (besides ribbons) and not unilamellar vesicles/tubes (besides ribbons), as described for the DOPE mixtures above. The GAP fit (Fig. 5M and N) results in dB = (59.3 ± 1.4) Å. After addition of DNA, the diffraction pattern of the Lcα phase occurs, and this fits perfectly with the observations made in TEM (Fig. 7C and D). Additionally, the signal of the Lα phase overlaps with a broad reflex, which is comparable to the reflex observed for the lipid mixture without DNA (see also Fig. S10, ESI‡), which indicated a hint of lipid blends without bound DNA. The Lcα signals result in a lamellar repeat distance d = 67.6 Å and a DNA rod distance of dDNA = 42.8 Å. The WAXS data (Fig. 5F) of the OH4/DPPC 1/1 (n/n) dispersion show a Bragg peak at q = 15.16 nm−1. The lipoplexes at N/P 4 show a comparable diffraction pattern (q = 15.16 nm−1). Therefore, a hexagonal chain lattice is assumed, as described for OH4 alone.
The TH4/DPPC 1/1 (n/n) mixture shows broad twisted ribbons and discs as dominant assemblies in the absence of DNA (Fig. 8A and B). Also, sheet-like layers are observed (Fig. S9, ESI‡). The negatively stained sample of TH4/DPPC 1/1 (n/n) lipoplexes shows larger lipoplex structures compared to the lipoplexes of the other lipid mixtures presented in this study, but lamellar ordering is still present (Fig. 8C, white triangles).
The SAXS pattern of TH4/DPPC 1/1 (n/n) without DNA exhibits a broad reflex comparable to the pattern of OH4/DPPC 1/1 (n/n) (compare Fig. 5E with Fig. 5G). The GAP fit results in dB = (53.9 ± 2.9) Å (Fig. 5O and P). The broad reflex also appears in the SAXS pattern of the TH4/DPPC 1/1 (n/n) mixture with DNA (see also Fig. S10, ESI‡). Additionally, two reflexes appear at q = 0.85 nm−1 and q = 1.06 nm−1. Due to the lamellar structure found in TEM images of TH4/DPPC 1/1 (n/n) lipoplexes (Fig. 8C), it is justified to assume the Lcα phase as the origin of these signals, but because of the missing second reflexes of the lamellar repeat distance, there is a certain degree of uncertainty. The lamellar repeat distance of the lipoplexes is d = 73.9 Å and the DNA rod distance is rather large, with dDNA = 59.4 Å. The detected WAXS patterns of the TH4/DPPC mixture in the presence and absence of DNA are comparable (Fig. 5H) and fit with the WAXS pattern of pure TH4 at pH 6.5 (Fig. 2F).
4 Discussion
The aggregation behaviour of the four lipid mixtures without DNA is quite different and gives evidence for characteristic factors determining an effective transfection system. It is obvious that the effective lipid blend, OH4/DOPE, is able to form liposomes (cryoTEM, Fig. 4A and B). This finding supports the thesis, which was established in earlier research, that the ability to form liposomes is directly connected with effective polynucleotide transfer.16 Also, the assumption of only partial lipid mixing of the OH4/DOPE composition16 is confirmed by the cryoTEM investigations, which show ribbons and sheets besides vesicles. We assume that the vesicles result from a mixture of OH4 and DOPE. In this mixed system, the positive curvature of OH4 and the negative curvature of DOPE result in a stable bilayer-forming system that forms liposomes.49 The alkyl chains of the liposome-forming lipid mixture seem to be in the fluid-crystalline phase state. This is supported by the DSC curve of the lipid mixture, which shows only a very broad and weak phase-transition peak (Fig. S11, ESI‡). The WAXS signals seem to result from an OH4-rich phase or phase-separated OH4, which is also responsible for the formation of ribbons and sheets. Evidence for this assumption is provided by the comparable WAXS patterns of the OH4/DOPE mixture and of OH4 alone (Table 1), with a slightly increased q11 value for the mixture. The formation of ribbons by amphiphiles with lysine or other amino acids in the head group is documented in the literature.50,51 This is also true for mixtures of lysine-containing lipids with DOPE.47 The sheets and ribbons are stabilized by H-bonds (see the above presented IRRAS and WAXS data for OH4). Unfortunately, the WAXS data for the OH4/DOPE are too weak to detect the reflex of the H-bond network. The OH4/DPPC mixture also forms the same kind of ribbons and sheets (besides discoidal assemblies), indicating that phase-separated OH4 is responsible for these assemblies. In this case, the WAXS signal is more intense, but we suggest that the reflex of the OH4-rich phase, and that of a separated phase of a DPPC/OH4 mixture, overlap (Fig. S12, ESI‡). IRRAS data for OH4/DOPE 1/1 as well as OH4/DPPC 1/1 give evidence of phase-separated OH4 in an H-bond network, supporting the thesis of H-bond stabilization of the ribbons and sheets (Fig. S13, ESI‡). The mixtures of TH4 with DOPE as well as DPPC also exhibit ribbons and sheets. In this case, the WAXS data for the mixtures (Fig. 5D and H) also give evidence that TH4-rich phases are responsible for these assemblies, because the patterns are comparable with that of pure TH4 (Fig. 2F and Table 1). Furthermore, the WAXS data (Table 1) as well as the IRRAS data (Fig. S13, ESI‡) of TH4/DOPE and TH4/DPPC mixtures, support the assumption that H-bonds stabilize the ribbons and sheets formed from TH4. The question arose why both cationic lipids form ribbons and sheets in a mixture with phospholipids, while OH4 alone forms a sponge-like structure and TH4 alone forms only sheets. The incorporation of the phospholipids in these assemblies provides an explanation. Whether the ribbons are long-time stable or metastable, and whether they form sheets by fusion or lipid exchange, was not investigated in this study.| OH4 | TH4 | OH4/DOPE | OH4/DPPC | TH4/DOPE | TH4/DPPC | |
|---|---|---|---|---|---|---|
| a Only detected in lipoplex dispersion. b Two q11 values because of two chain lattices, resulting in two peaks, see Fig. S12, ESI. c From orthorhombic lattice or Cα–Cα-spacing. | ||||||
| Chain lattice | Hexagonal | Hexagonal/orthorhom. | Hexagonal | Hexagonalb | Hexagonal/orthorhom. | Hexagonalb/orthorhom. |
| q 11 = 15.25 nm−1 | q 11 = 15.35 nm−1 | q 11 = 15.39 nm−1 | q 11 = 15.16 nm−1 | q 11 = 15.39 nm−1 | q 11 = 15.21 nm−1 | |
| q 11 = 15.45 nm−1 | q 11 = 15.36 nm−1 | |||||
| q H-bond | 13.04 nm−1 | 12.98 nm−1 | — | — | 13.05 nm−1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
13.02 nm−1 |
| Additional peakc | — | q = 16.50 nm−1 | — | — | q = 16.63 nm−1 | q = 16.65 nm−1 |
The two DOPE-containing mixtures are able to form liposomes. As mentioned above, OH4/DOPE 1/1 mixtures form unilamellar liposomes with a size of 30 nm to 200 nm. In contrast, TH4/DOPE 1/1 builds liposomes on the μm scale, which show evidence of lateral de-mixing processes, resulting in budding of smaller vesicles, tube formation and the formation of constricted tubes (Fig. 6A–C). These structures are the results of DOPE-rich regions with negative curvature effects and TH4-rich regions with positive curvature effects in the lipid membrane. Such a phenomenon, known as GUV pearling, is also described in the literature for other DOPE-containing lipid mixtures.52 Keeping in mind the fact that OH4/DOPE is a much more effective gene transfer system than TH4/DOPE, we update the assumption of the positive effect of liposome formation to: malonic acid diamide blends that form stable liposomes are efficient gene carriers.
DPPC in mixtures with OH4, as well as TH4 tends to form lipid discs (bicelles), as well as ribbons and sheets. As discussed before, we prefer the model of a mixture of a cationic lipid with DMPC.53 In this case, the body of the discs is formed by the phospholipids or a phospholipid-rich lipid mixture and the rim of the disc is stabilized by the cationic lipid (Fig. 9B). Such stabilization of bilayer discs formed by phosphatidylcholines is also discussed in the literature with respect to amphiphiles with bulky head groups (PEG-lipids and T-shaped amphiphiles) and proteins.54–56 There is clear evidence for the advantages of this model: (i) DPPC has no curvature effect, while the two cationic lipids induce a positive membrane curvature; (ii) Denisov et al. described DPPC discs with thicknesses of 55 Å and 59 Å, which are comparable with the dB values determined in this study (Table 2);55 (iii) the q11 WAXS signals consist of two signals from different chain lattices (Table 1). Deconvolution results in two peaks for both mixtures (OH4/DPPC, TH4/DPPC) (Fig. S12, ESI‡). None of the deconvoluted peaks are comparable to the WAXS signal of pure DPPC in the Lβ′ phase (Fig. S12, ESI‡), which indicates the distinct incorporation of the cationic lipid into the DPPC bilayer. This assumption perfectly fits with the DSC curves of the OH4/DPPC and TH4/DPPC mixtures (Fig. S11, ESI‡), which are not comparable with those of pure DPPC,16 and with the phase separation processes published earlier.16 This leads to the assumption that the body of the bicelles is formed by a DPPC-rich phase instead of pure DPPC (model Fig. 9B).
| Parameters | OH4/DOPE | OH4/DPPC | TH4/DOPE | TH4/DPPC | |
|---|---|---|---|---|---|
| −DNA | d B [Å] | 55.6 ± 1.5 | 59.3 ± 1.4 | 55.2 ± 1.6 | 53.9 ± 2.9 |
| d B (mixture)–dB (cationic lipid) [Å] | 8.1 ± 7.1 | 11.6 ± 2.7 | 7.5 ± 2.8 | 6.2 ± 3.7 | |
| +DNA | d [Å] | 69.0 | 67.6 | 69.8 | 73.9 |
| d–dB [Å] | 13.4 ± 1.5 | 8.3 ± 1.4 | 14.6 ± 1.6 | 20.0 ± 2.9 | |
| d DNA [Å] | 46.2 | 42.8 | 39.5 | 59.4 |
All four investigated lipid mixtures, the effective as well as the ineffective DNA delivery systems, form lipoplexes with the Lcα structure (model Fig. 9C). Nevertheless, there are clear differences between the different lipid mixtures in complexing DNA, but distinct connections to the DNA transfer efficiency can hardly be made. Firstly, there are hints of de-mixing processes in a DNA-containing and a DNA-free phase. OH4/DPPC shows clear de-mixing in negative-stain TEM and cryoTEM experiments, where discs and ribbons exist besides lipoplexes. Unfortunately, the coexistence of lipoplexes and ribbons can only be detected by cryoTEM, a technique that induces beam damage in large lipoplex structures. Secondly, we cannot exclude de-mixing processes in a DNA-containing lipoplex and DNA-free structures for the other investigated lipid mixtures. The SAXS curves for the lipoplexes of DPPC-containing mixtures, as well as for TH4/DOPE, also give evidence for additional structures. Also, the fact that all lipoplex structures are investigated at N/P = 4, an N/P ratio above the complex formation endpoint between DNA and the cationic lipid mixture,14 underlines the assumption of DNA-free structures in all investigated lipoplex formulations. Different types of lipoplexes were not found in one lipoplex formulation. Thus, the lipoplexes consisting of lipid ribbons stacked together by DNA, described by Junquera and coworkers, were not observed,47 although we observed ribbons in all lipid formulations.
We will now focus on the data for the Lcα SAXS pattern. The bilayer repeat distance (d) is comparable for all lipoplexes, except for the TH4/DPPC lipoplexes (ineffective DNA delivery system), which exhibit a higher value (Table 2). The difference between the lipid bilayer thickness of the lipid mixture without DNA, determined by the GAP fit (dB) and d, yields the thickness of the DNA-containing sublayer. The diameter of the DNA is around 20 Å.57 This value is only observed in the TH4/DPPC blend. The lipoplex formulations of the other three mixtures yield a DNA-containing layer thickness considerably below 2 nm. This can be explained by three models: (i) the DNA strands partially penetrate into the head group layers of the lipid bilayer; (ii) the interaction with the DNA changes the property of the lipid bilayer, resulting in a smaller bilayer compared to that obtained in the lipid blends in the absence of DNA, (iii) the thickness of the DNA-containing sub-layer can fluctuate between 2 nm at the location of the DNA strands, with smaller values in the DNA-free regions (Fig. 9C), resulting in a smaller averaged DNA-containing layer thickness. Nevertheless, this finding can be a hint of weaker DNA binding in TH4/DPPC lipoplexes, compared to the other 3 investigated systems.
Finally, the dDNA value is dependent on the charge density of the lipid bilayer of the complex.58,59 An increase in charge density results in a decrease in dDNA. Due to the DNA diameter of 20 Å, the smallest possible dDNA is in the same range, but keeping in mind the electrostatic repulsion of charged DNA molecules, the smallest reachable value has to be higher. The highly effective composite, OH4/DOPE, exhibits a dDNA value of 46.2 Å. Safinya and co-workers postulated a bell-shaped curve for the transfection efficiency as a function of the charge density in the case of Lcα lipoplexes.60,61 The weak transfecting mixtures, OH4/DPPC and TH4/DOPE, show slightly lower dDNA values, indicating a higher charge density of the complexing lipid composite, leading to a less effective DNA release from the lipoplex. The ineffective mixture, TH4/DPPC, exhibits quite a large dDNA value, indicating DNA binding that is too weak. A different quality of DNA binding in the investigated lipoplexes can be also seen in DSC experiments (Fig. S11, ESI‡). The melting of the DNA double strand into single strands occurs between 55 °C and 75 °C. All four lipoplex formulations show a transition at lower temperatures, indicating an earlier DNA melting. This indicates a DNA double-strand destabilization. The transition enthalpies of the low-temperature melting transitions of DNA in the lipoplex formulations are very low, compared to the transition enthalpy of pure DNA, except for the enthalpy of DNA denaturation in the TH4/DPPC blend. Furthermore, all lipoplex formulations show an increase in the heat capacity at the end of the curve, indicating a high temperature transition, which can be assigned to DNA melting of compacted DNA in lipoplexes.62 Taken together, the DSC data support the fact that TH4/DPPC is not able to compact the DNA in an efficient manner.
The mixing behaviour of the cationic lipid and the co-lipid affects the geometry and the charge density of lipid aggregates. For instance, the cationic lipid-rich ribbons have a very high charge density, while the vesicles have a lower charge density, because the neutral co-lipid is incorporated in the bilayer. The discs have a low charge density, except at the cationic lipid-rich rim, where the charge density is high. Nevertheless, the lipoplex formation leads to lipid rearrangement (Lcα formation); the character of the preformed types of aggregates seems to affect the quality and strength of DNA binding inside the lipoplex.
Finally, it should be noted that this study characterizes lipid structures, which are of potential interest for other applications as well as gene therapy. For instance, the sponge-like structure of OH4 may be of interest for peptides used in vaccination. Also, the cationic lipid discs can be potentially used as carrier systems for anionic drugs.
5 Conclusion
The structural investigations demonstrate that slight changes in the structure of cationic lipids, resulting from introducing a tetradecyl chain instead of an oleyl chain, have pronounced effects on the aggregation behaviour as well as on the mixing behaviour with phospholipids. While OH4 forms sponge-like networks stabilized by hydrogen bonds, TH4 forms hydrogen-bond-stabilized sheets. Both lipids do not ideally mix with DOPE or DPPC, resulting in ribbon-like structures formed from a cationic lipid-rich mixture beside the main structure, which is formed from a binary cationic lipid/phospholipid mixture. OH4/DOPE forms stable unilamellar vesicles (effective gene transfer), while TH4/DOPE forms vesicles that show instability due to lateral de-mixing processes. The OH4/DPPC and TH4/DPPC mixtures result in discoidal structures. Despite the differences in the structures of the lipid aggregates, all four investigated binary mixtures result in an Lcα lipoplex structure after complexation with DNA, but the quality and character of the DNA binding differs in all four lipoplexes, as demonstrated by the different intensity of the SAXS signals, the thickness of the DNA-containing sub-layer, the distance between the aligned DNA rods, and DSC experiments. The character of the formed lipid assemblies, as well as the quality of binding between the lipid mixtures and the DNA, affects the transfection efficiency.Acknowledgements
We are grateful to Dr Simon Drescher (Martin Luther University, Halle-Wittenberg) for the preparation of the negatively stained TEM samples. We thank the BESSY II facility for the beam time at the MySPOT beamline and Dr Ivo Zizak for support during the X-ray measurements. Furthermore, we thank ESRF (Grenoble, France) for attributed beam time at the High Brilliance Beamline ID02 and Dr Gudrun Lotze for the excellent support. We thank Georg Pabst (University of Graz, Austria) for providing the GAP program.References
- M. L. Maeder and C. A. Gersbach, Mol. Ther., 2016, 24, 430–446 CAS.
- L. Naldini, Nature, 2015, 526, 351–360 CrossRef CAS PubMed.
- C. Sheridan, Nat. Biotechnol., 2011, 29, 121–128 CrossRef CAS PubMed.
- E. Wagner, Acc. Chem. Res., 2012, 45, 1005–1013 CrossRef CAS PubMed.
- S. L. Ginn, I. E. Alexander, M. L. Edelstein, M. R. Abedi and J. Wixon, J. Gene Med., 2013, 15, 65–77 CrossRef CAS PubMed.
- P. L. Felgner, T. R. Gadek, M. Holm, R. Roman, H. W. Chan, M. Wenz, J. P. Northrop, G. M. Ringold and M. Danielsen, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 7413–7417 CrossRef CAS.
- C.-L. Chan, K. K. Ewert, R. N. Majzoub, Y.-K. Hwu, K. S. Liang, C. Leal and C. R. Safinya, J. Gene Med., 2014, 16, 84–96 CrossRef CAS PubMed.
- M. B. de Jesus and I. S. Zuhorn, J. Controlled Release, 2015, 201, 1–13 CrossRef CAS PubMed.
- K. Ewert, A. Ahmad, H. M. Evans and C. R. Safinya, Expert Opin. Biol. Ther., 2005, 5, 33–53 CrossRef CAS PubMed.
- E. Kai, L. S. Nelle, A. Ayesha, M. E. Heather, J. L. Alison, E. S. Charles and R. S. Cyrus, Curr. Med. Chem., 2004, 11, 133–149 CrossRef.
- R. Koynova, B. Tenchov, L. Wang and R. C. MacDonald, Mol. Pharmaceutics, 2009, 6, 951–958 CrossRef CAS PubMed.
- R. Koynova and B. Tenchov, Soft Matter, 2009, 5, 3187–3200 RSC.
- B. G. Tenchov, L. Wang, R. Koynova and R. C. MacDonald, Biochim. Biophys. Acta, Biomembr., 2008, 1778, 2405–2412 CrossRef CAS PubMed.
- C. H. Jones, C.-K. Chen, A. Ravikrishnan, S. Rane and B. A. Pfeifer, Mol. Pharmaceutics, 2013, 10, 4082–4098 CrossRef CAS PubMed.
- C. Wölk, S. Drescher, A. Meister, A. Blume, A. Langner and B. Dobner, Chem. – Eur. J., 2013, 19, 12824–12838 CrossRef PubMed.
- C. Janich, C. Wölk, S. Taßler, S. Drescher, A. Meister, G. Brezesinski, B. Dobner and A. Langner, Eur. J. Lipid Sci. Technol., 2014, 116, 1184–1194 CrossRef CAS.
- C. Janich, C. Wölk, F. Erdmann, T. Groth, G. Brezesinski, B. Dobner and A. Langner, J. Controlled Release, 2015, 220(Part A), 295–307 CrossRef CAS PubMed.
- C. Stefaniu, G. Brezesinski and H. Möhwald, Adv. Colloid Interface Sci., 2014, 208, 197–213 CrossRef CAS PubMed.
- J. D. Hartgerink, J. R. Granja, R. A. Milligan and M. R. Ghadiri, J. Am. Chem. Soc., 1996, 118, 43–50 CrossRef CAS.
- G. Pabst, R. Koschuch, B. Pozo-Navas, M. Rappolt, K. Lohner and P. Laggner, J. Appl. Crystallogr., 2003, 36, 1378–1388 CrossRef CAS.
- G. Pabst, M. Rappolt, H. Amenitsch and P. Laggner, Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top., 2000, 62, 4000–4009 CrossRef CAS.
- I. Winter, G. Pabst, M. Rappolt and K. Lohner, Chem. Phys. Lipids, 2001, 112, 137–150 CrossRef CAS PubMed.
- A. H. Muenter, J. Hentschel, H. G. Börner and G. Brezesinski, Langmuir, 2008, 24, 3306–3316 CrossRef CAS PubMed.
- B. Angelov, A. Angelova, R. Mutafchieva, S. Lesieur, U. Vainio, V. M. Garamus, G. V. Jensen and J. S. Pedersen, Phys. Chem. Chem. Phys., 2011, 13, 3073–3081 RSC.
- L. C. Serpell, Biochim. Biophys. Acta, Mol. Basis Dis., 2000, 1502, 16–30 CrossRef CAS.
- J. T. Nguyen, H. Inouye, M. A. Baldwin, R. J. Fletterick, F. E. Cohen, S. B. Prusiner and D. A. Kirschner, J. Mol. Biol., 1995, 252, 412–422 CrossRef CAS PubMed.
- M. Dittrich, M. Heinze, C. Wölk, S. S. Funari, B. Dobner, H. Möhwald and G. Brezesinski, ChemPhysChem, 2011, 12, 2328–2337 CrossRef CAS PubMed.
- G. Brezesinski, B. Dobner, C. Stefaniu and D. Vollhardt, J. Phys. Chem. C, 2011, 115, 8206–8213 CAS.
- I. W. Hamley, A. Dehsorkhi, V. Castelletto, M. N. M. Walter, C. J. Connon, M. Reza and J. Ruokolainen, Langmuir, 2015, 31, 4490–4495 CrossRef CAS PubMed.
- I. Kuzmenko, V. M. Kaganer and L. Leiserowitz, Langmuir, 1998, 14, 3882–3888 CrossRef CAS.
- l. A. Fedotenko, C. Stefaniu, G. Brezesinski and A. Zumbuehl, Langmuir, 2013, 29, 9428–9435 CrossRef CAS PubMed.
- R. S. Clegg and J. E. Hutchison, Langmuir, 1996, 12, 5239–5243 CrossRef CAS.
- X. Du, W. Miao and Y. Liang, J. Phys. Chem. B, 2005, 109, 7428–7434 CrossRef CAS PubMed.
- T. D. Andreeva, J. G. Petrov, G. Brezesinski and H. Moehwald, Langmuir, 2008, 24, 8001–8007 CrossRef CAS PubMed.
- G. Brezesinski, B. Dobner, C. Stefaniu and D. Vollhardt, Langmuir, 2011, 27, 5386–5392 CrossRef CAS PubMed.
- A. Blume and A. Kerth, Biochim. Biophys. Acta, Biomembr., 2013, 1828, 2294–2305 CrossRef CAS.
- R. Mendelsohn, G. Mao and C. R. Flach, Biochim. Biophys. Acta, Biomembr., 2010, 1798, 788–800 CrossRef CAS PubMed.
- A. Zumbuehl, B. Dobner and G. Brezesinski, Curr. Opin. Colloid Interface Sci., 2014, 19, 17–24 CrossRef CAS.
- A. Weinberger, R. Tanasescu, C. Stefaniu, l. A. Fedotenko, F. Favarger, T. Ishikawa, G. Brezesinski, C. M. Marques and A. Zumbuehl, Langmuir, 2015, 31, 1879–1884 CrossRef CAS PubMed.
- P. W. M. Van Dijck, Biochim. Biophys. Acta, Biomembr., 1979, 555, 89–101 CrossRef CAS.
- P. R. Cullis, P. W. M. Van Dijck, B. De Kruijff and J. De Gier, Biochim. Biophys. Acta, Biomembr., 1978, 513, 21–30 CrossRef CAS.
- R. N. A. H. Lewis, N. Mak and R. N. McElhaney, Biochemistry, 1987, 26, 6118–6126 CrossRef CAS PubMed.
- J. Ulminus, H. Wennerstrom, G. Lindblom and G. Arvidson, Biochemistry, 1977, 16, 5742–5745 CrossRef CAS.
- B. Ma, S. Zhang, H. Jiang, B. Zhao and H. Lv, J. Controlled Release, 2007, 123, 184–194 CrossRef CAS PubMed.
- I. Koltover, T. Salditt, J. O. Rädler and C. R. Safinya, Science, 1998, 281, 78–81 CrossRef CAS PubMed.
- S. Mochizuki, N. Kanegae, K. Nishina, Y. Kamikawa, K. Koiwai, H. Masunaga and K. Sakurai, Biochim. Biophys. Acta, Biomembr., 2013, 1828, 412–418 CrossRef CAS.
- A. L. Barran-Berdon, M. Munoz-Ubeda, C. Aicart-Ramos, L. Perez, M.-R. Infante, P. Castro-Hartmann, A. Martin-Molina, E. Aicart and E. Junquera, Soft Matter, 2012, 8, 7368–7380 RSC.
- J. O. Rädler, I. Koltover, T. Salditt and C. R. Safinya, Science, 1997, 275, 810–814 CrossRef.
- H. J. Risselada and S. J. Marrink, Phys. Chem. Chem. Phys., 2009, 11, 2056–2067 RSC.
- L. Ziserman, H.-Y. Lee, S. R. Raghavan, A. Mor and D. Danino, J. Am. Chem. Soc., 2011, 133, 2511–2517 CrossRef CAS PubMed.
- H. Cui, A. G. Cheetham, E. T. Pashuck and S. I. Stupp, J. Am. Chem. Soc., 2014, 136, 12461–12468 CrossRef CAS PubMed.
- M. Kang, G. Huang and C. Leal, Soft Matter, 2014, 10, 8846–8854 RSC.
- C. Wölk, C. Janich, A. Meister, S. Drescher, A. Langner, G. Brezesinski and U. Bakowsky, Bioconjugate Chem., 2015, 26, 2461–2473 CrossRef PubMed.
- P. Scholtysek, A. Achilles, C.-V. Hoffmann, B.-D. Lechner, A. Meister, C. Tschierske, K. Saalwächter, K. Edwards and A. Blume, J. Phys. Chem. B, 2012, 116, 4871–4878 CrossRef CAS PubMed.
- I. G. Denisov, Y. V. Grinkova, A. A. Lazarides and S. G. Sligar, J. Am. Chem. Soc., 2004, 126, 3477–3487 CrossRef CAS PubMed.
- M. M. Zetterberg, K. Reijmar, M. Pränting, Å. Engström, D. I. Andersson and K. Edwards, J. Controlled Release, 2011, 156, 323–328 CrossRef CAS PubMed.
- R. Podgornik, D. C. Rau and V. A. Parsegian, Macromolecules, 1989, 22, 1780–1786 CrossRef CAS.
- G. Caracciolo, D. Pozzi, H. Amenitsch and R. Caminiti, Langmuir, 2006, 22, 4267–4273 CrossRef CAS PubMed.
- O. Farago and N. Grønbech-Jensen, Biophys. J., 2007, 92, 3228–3240 CrossRef CAS PubMed.
- A. Ahmad, H. M. Evans, K. Ewert, C. X. George, C. E. Samuel and C. R. Safinya, J. Gene Med., 2005, 7, 739–748 CrossRef CAS PubMed.
- C. R. Safinya, K. K. Ewert, R. N. Majzoub and C. Leal, New J. Chem., 2014, 38, 5164–5172 RSC.
- C. Wölk, D. Pawlowska, S. Drescher, A. Auerswald, A. Meister, G. Hause, A. Blume, A. Langner, G. Brezesinski and B. Dobner, Langmuir, 2014, 30, 4905–4915 CrossRef PubMed.
Footnotes |
| † This article is dedicated to Professor Andreas Langner on occasion of his 60th birthday. We are very grateful for the allocation of the innovative research topic. |
| ‡ Electronic supplementary information (ESI) available: Additional IRRAS, TEM, and X-ray data, as well as differential scanning calorimetry and atomic force microscopy experiments. See DOI: 10.1039/c6sm00807k |
| This journal is © The Royal Society of Chemistry 2016 |