Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Structure of the H-NS–DNA nucleoprotein complex†
Johan R. C.
van der Maarel
*a,
Durgarao
Guttula
a,
Véronique
Arluison
bc,
Stefan U.
Egelhaaf
d,
Isabelle
Grillo
e and
V. Trevor
Forsyth
ef
aDepartment of Physics, National University of Singapore, Singapore 117542, Singapore. E-mail: johanmaarel@gmail.com; Fax: +65 6777612; Tel: +65 65164396
bLaboratoire Léon Brillouin UMR 12 CEA, CNRS, Université Paris Saclay, CEA–Saclay, Gif sur Yvette Cedex 91191, France
cUniversité Paris Diderot, Sorbonne Paris Cité, 75013 Paris, France
dHeinrich-Heine University, Condensed Matter Physics Laboratory, Düsseldorf, 40225, Germany
eInstitut Laue-Langevin, 38042 Grenoble, France
fKeele University, Faculty of Natural Sciences, Staffordshire, ST5 5BG, UK
First published on 9th March 2016
Abstract
Nucleoid associated proteins (NAPs) play a key role in the compaction and expression of the prokaryotic genome. Here we report the organisation of a major NAP, the protein H-NS on a double stranded DNA fragment. For this purpose we have carried out a small angle neutron scattering study in conjunction with contrast variation to obtain the contributions to the scattering (structure factors) from DNA and H-NS. The H-NS structure factor agrees with a heterogeneous, two-state binding model with sections of the DNA duplex surrounded by protein and other sections having protein bound to the major groove. In the presence of magnesium chloride, we observed a structural rearrangement through a decrease in cross-sectional diameter of the nucleoprotein complex and an increase in fraction of major groove bound H-NS. The two observed binding modes and their modulation by magnesium ions provide a structural basis for H-NS-mediated genome organisation and expression regulation.
1 Introduction
The prokaryotic genome is highly compacted in the nucleoid, despite the lack of scaffolding in terms of lipid membranes and/or chromatin organization.1 Although much work has been done to elucidate the mechanisms involved in stabilising the compacted state, the structural arrangement of the condensing agents near DNA is not clear. In previous small angle neutron scattering (SANS) works, we have measured the distribution of ions, including polyamines, around DNA.2–6 More recently, we have investigated the structure of the bacterial protein Hfq bound to DNA.7 Here, we describe the structure of the complex formed by the generic nucleoid associated protein H-NS (histone-like nucleoid structuring protein, Mw = 15.6 kDa) with double-stranded DNA. H-NS plays a global role in gene regulation and represses hundreds of genes, most of which are involved in the adaptation to stress, virulence and chemotaxis.8 It exists as a dimer as well as higher oligomeric forms and binds DNA with a preference for curved or AT-rich sequences. These sequences serve as nucleation sites for oligomerization of the protein along the duplex; thus covering regions as long as 1.5 kbp in vivo.9 H-NS binding results in an increase in bending rigidity and/or bridging of distant, like-charged segments of the DNA molecule by protein-mediated attraction.10,11 The relative importance of bridging and stiffening depends on buffer composition and, in particular, the presence of divalent cations (Mg2+).12,13 The formation of the nucleoprotein complex has been proposed to be the structural basis for H-NS-mediated gene silencing.14–17Detailed structural information on the binding of H-NS to DNA is scarce. Two distinguishable H-NS binding states have been identified, depending on the interaction of specific and nonspecific DNA target sites.18 H-NS specifically binds to the minor groove of double-stranded DNA with a short C-terminal loop.19 Nonspecific binding is thought to be predominantly controlled by electrostatics and is much more prone to variation in ionic strength. Here, we describe a SANS study of H-NS complexed to rod-like DNA fragments (contour length 54 nm) in solution with monovalent and divalent salts. The contributions to the scattering (structure factors) from DNA and H-NS are obtained using solvent contrast variation. The H-NS to DNA base-pair ratio was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6, so that the DNA fragments are almost fully covered with protein. Information on the arrangement of H-NS about B-form DNA is obtained by comparison of the H-NS structure factor with coarse-grained model calculations involving the radial distribution in amino acid density. Key structural features of the nucleoprotein complex are derived, including the cross-sectional radius of gyration and the extent to which H-NS penetrates the grooves. In particular, the effect of magnesium ions on the structure of the complex is explored. The structural arrangement of bound H-NS is discussed in the context of H-NS-mediated genome organisation and gene expression regulation.
6, so that the DNA fragments are almost fully covered with protein. Information on the arrangement of H-NS about B-form DNA is obtained by comparison of the H-NS structure factor with coarse-grained model calculations involving the radial distribution in amino acid density. Key structural features of the nucleoprotein complex are derived, including the cross-sectional radius of gyration and the extent to which H-NS penetrates the grooves. In particular, the effect of magnesium ions on the structure of the complex is explored. The structural arrangement of bound H-NS is discussed in the context of H-NS-mediated genome organisation and gene expression regulation.
2 Materials and methods
2.1 Sample preparation
DNA fragments (150 bp) were obtained by micrococcal digestion of calf thymus chromatin.20 His-tagged Escherichia coli H-NS was purified from over-expressing BL21(DE3)/pLATE 31-hns cells. The pLATE 31-hns expression vector was constructed by the ligation independent cloning method (Thermo Fisher). As the His-tag potentially modifies the properties of H-NS, a Tobacco Etch Virus (TEV) protease site ENLYFQG was inserted between the C-terminal His-tag and H-NS sequence. The oligonucleotides used for cloning were AGAAGGAGATATAACTATGAGCGAAGCACTTAAAATTCTGAAC (for) and GTGGTGGTGATGGTGATGGCCCTGGAAGTACAGGTTCTCTTGCTTGATCAGGAAATCGT (rev). Cells from post-induction cultures were resuspended in 20 mM Tris-HCl pH 7.5, 0.5 M NaCl, 10% (v/v) glycerol and a protease inhibitor (Sigma) at 277 K. The suspension was sonicated and the lysed cells were cleared by centrifugation at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min. DNase I (40 g L−1) and RNase A (30 g L−1) were added to the cleared lysate at 303 K. The solution was then applied to a Ni2+–NTA column (GE Healthcare). The resin was washed with 20 mM Tris-HCl, pH 7.8, 0.3 M NaCl, 20 mM imidazole and the protein was eluted with a gradient of imidazole (20–500 mM). TEV digestion was carried out according to the manufacturer's instructions (Thermo Fisher). After digestion, fragmented and non-digested H-NS were removed with Ni2+–NTA magnetic beads (GE Healthcare). Several rounds of purification were performed to obtain the required amount of 50 mg H-NS. For gel electrophoresis characterisation of DNA and H-NS see ESI,† Fig. S1 and S2, respectively, online.
000g for 30 min. DNase I (40 g L−1) and RNase A (30 g L−1) were added to the cleared lysate at 303 K. The solution was then applied to a Ni2+–NTA column (GE Healthcare). The resin was washed with 20 mM Tris-HCl, pH 7.8, 0.3 M NaCl, 20 mM imidazole and the protein was eluted with a gradient of imidazole (20–500 mM). TEV digestion was carried out according to the manufacturer's instructions (Thermo Fisher). After digestion, fragmented and non-digested H-NS were removed with Ni2+–NTA magnetic beads (GE Healthcare). Several rounds of purification were performed to obtain the required amount of 50 mg H-NS. For gel electrophoresis characterisation of DNA and H-NS see ESI,† Fig. S1 and S2, respectively, online.
Two sets of samples with 3.2 g of DNA/L and 12.7 g of H-NS/L (H-NS to bp ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) in Tris-HCl (T) buffer with 100 mM KCl and T-buffer with 100 mM KCl and 10 mM MgCl2, respectively, were prepared. The Tris-HCl concentration is 10 mM Tris adjusted with HCl to pH 7.5 (i.e., 8.1 mM Tris-Cl and 1.9 mM Tris). For contrast variation, each sample set was prepared in 0%, 43% (H-NS-matched), 64% (DNA-matched), and 99% D2O. Reference solvent samples with matching H2O/D2O composition were also prepared. All manipulations involving D2O were performed under a flow of Argon in order to minimise exchange with atmospheric water. The solvent compositions were determined by weight and checked by the values for transmission. Standard quartz cuvettes with 0.1 cm (for samples in pure H2O) or 0.2 cm path length were used.
6) in Tris-HCl (T) buffer with 100 mM KCl and T-buffer with 100 mM KCl and 10 mM MgCl2, respectively, were prepared. The Tris-HCl concentration is 10 mM Tris adjusted with HCl to pH 7.5 (i.e., 8.1 mM Tris-Cl and 1.9 mM Tris). For contrast variation, each sample set was prepared in 0%, 43% (H-NS-matched), 64% (DNA-matched), and 99% D2O. Reference solvent samples with matching H2O/D2O composition were also prepared. All manipulations involving D2O were performed under a flow of Argon in order to minimise exchange with atmospheric water. The solvent compositions were determined by weight and checked by the values for transmission. Standard quartz cuvettes with 0.1 cm (for samples in pure H2O) or 0.2 cm path length were used.
2.2 Small angle neutron scattering
SANS experiments were carried out using the D11 diffractometer at the Institut Laue–Langevin, Grenoble. A wavelength of 0.6 nm with a 10% spread was selected and the sample-detector distances were 1.1 and 13.5 m, respectively. The total counting times for all detector settings was approximately 2 h per sample. Data reduction allowed subtraction of background scattering, sample transmission and detector pixel efficiency. The efficiencies of the detector pixels were determined using the scattering of H2O. Absolute intensities were obtained with reference to pure water, and the scattering of the sample cell with solvent at the same isotopic composition was subtracted. The sample temperature was 298 K.2.3 Contrast variation
We have used contrast variation to match or highlight the nucleic acid and protein components of the nucleoprotein complex. The scattering contrast is hence a key experimental parameter. The scattering length contrast of the nucleotides (i = n) and amino acids (i = a) with respect to solvent (water) is given by![[b with combining macron]](https://www.rsc.org/images/entities/i_char_0062_0304.gif) i = bi − bs i = bi − bs![[v with combining macron]](https://www.rsc.org/images/entities/i_char_0076_0304.gif) i/ i/![[v with combining macron]](https://www.rsc.org/images/entities/i_char_0076_0304.gif) s. s. | (1) |
![[v with combining macron]](https://www.rsc.org/images/entities/i_char_0076_0304.gif) i/
i/![[v with combining macron]](https://www.rsc.org/images/entities/i_char_0076_0304.gif) s. In a mixture of H2O and D2O, the scattering length of the solvent is
s. In a mixture of H2O and D2O, the scattering length of the solvent is| bs = xbD2O + (1 − x)bH2O, | (2) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) G
G![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C
C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) T
T![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5-methylcytosine = 0.28
5-methylcytosine = 0.28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.22
0.22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.21
0.21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.28
0.28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.01 and the amino acid sequence of H-NS (UniProt database, accession number P0ACF8). The results are shown in Table 1. Scattering length contrasts pertaining to the different H2O/D2O solvent compositions of our samples were calculated with eqn (1) and (2) using the parameters in Table 1 and are shown in Table 2.
0.01 and the amino acid sequence of H-NS (UniProt database, accession number P0ACF8). The results are shown in Table 1. Scattering length contrasts pertaining to the different H2O/D2O solvent compositions of our samples were calculated with eqn (1) and (2) using the parameters in Table 1 and are shown in Table 2.
![[v with combining macron]](https://www.rsc.org/images/entities/i_char_0076_0304.gif) and neutron scattering lengths b of the solutes and solvents given (x denotes the D2O mole or volume fraction)
and neutron scattering lengths b of the solutes and solvents given (x denotes the D2O mole or volume fraction)
![[v with combining macron]](https://www.rsc.org/images/entities/i_char_0076_0304.gif) (cm3 mole−1) (cm3 mole−1) |
b (10−12 cm) | |
|---|---|---|
| DNA | 172 | 9.77 + 2.02x |
| H-NS | 84 | 2.50 + 2.06x |
| H2O | 18 | −0.168 |
| D2O | 18 | 1.915 |
![[b with combining macron]](https://www.rsc.org/images/entities/i_char_0062_0304.gif) n and amino acid
n and amino acid ![[b with combining macron]](https://www.rsc.org/images/entities/i_char_0062_0304.gif) a scattering length contrasts in different H2O/D2O mixtures
a scattering length contrasts in different H2O/D2O mixtures
![[b with combining macron]](https://www.rsc.org/images/entities/i_char_0062_0304.gif) n (10−12 cm)
n (10−12 cm) |
![[b with combining macron]](https://www.rsc.org/images/entities/i_char_0062_0304.gif) a (10−12 cm)
a (10−12 cm) |
|
|---|---|---|
| H2O | 11.4 | 3.3 |
| 43% D2O | 3.7 | 0.0 |
| 64% D2O | −0.1 | −1.6 |
| 99% D2O | −6.3 | −4.3 |
2.4 From intensities to structure factors
In the analysis of the structure factors, the nucleotides and the amino acids are considered to be the elementary scattering units. For a solution of nucleoprotein complexes with density ρf, the coherent part of the solvent corrected scattered intensity is the sum of three partial structure factors describing the density correlations among the nucleotides and amino acidsI(q)/ρf = Nn2![[b with combining macron]](https://www.rsc.org/images/entities/i_char_0062_0304.gif) n2Sn(q) + 2NnNa n2Sn(q) + 2NnNa![[b with combining macron]](https://www.rsc.org/images/entities/i_char_0062_0304.gif) n n![[b with combining macron]](https://www.rsc.org/images/entities/i_char_0062_0304.gif) aSna(q) + Na2 aSna(q) + Na2![[b with combining macron]](https://www.rsc.org/images/entities/i_char_0062_0304.gif) a2Sa(q) a2Sa(q) | (3) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6, Na exceeds Nn by more than an order of magnitude. Unless the protein is exactly matched with
6, Na exceeds Nn by more than an order of magnitude. Unless the protein is exactly matched with ![[b with combining macron]](https://www.rsc.org/images/entities/i_char_0062_0304.gif) a = 0, the scattering is dominated by protein. The contribution from the small ions to the scattering at small angles is negligible. Momentum transfer q is defined by the wavelength λ of the radiation and scattering angle θ according to q = 4π/λ
a = 0, the scattering is dominated by protein. The contribution from the small ions to the scattering at small angles is negligible. Momentum transfer q is defined by the wavelength λ of the radiation and scattering angle θ according to q = 4π/λ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sin(θ/2). The partial structure factors Sij(q) with i, j = n, a (Sii is abbreviated as Si) are the spatial Fourier transforms of the nucleotide and amino acid density correlation functions and are normalised to unity at q = 0.
sin(θ/2). The partial structure factors Sij(q) with i, j = n, a (Sii is abbreviated as Si) are the spatial Fourier transforms of the nucleotide and amino acid density correlation functions and are normalised to unity at q = 0.
The nucleoprotein complex can be seen as a cylindrical object with a length L. Any possible ordering of protein in register with the phosphate moieties along the DNA molecule is beyond detection in scattering from solution given isotropic ensemble averaging. In the longitudinal direction (along the DNA molecule), the nucleotide and amino acid distributions are hence assumed to be uniform. In the perpendicular direction, away from the axis of the complex, the corresponding distributions are given by the radial profiles ρn(r) and ρa(r), respectively. In the present range of momentum transfer, the scattering is sensitive to correlations over distances of the order of the thickness of the complex and effects of finite contour length and flexibility are negligible. The partial structure factors can then be expressed as a product of a term related to the structure of an equivalent solution of complexes with vanishing cross-section and terms involving the radial profiles
| Sij(q) = S(q)ai(q)aj(q) | (4) |
 | (5) |
| Sn = πan2(q)/qL and Sa = πaa2(q)/qL (qL ≫ 1) | (6) |
3 Results
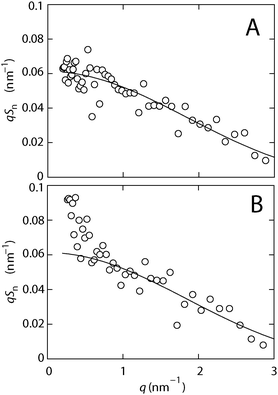
SANS experiments were carried out on a system of 150 bp DNA fragments and H-NS in 10 mM Tris-HCl (T) buffer with 100 mM KCl, and in T-buffer with 100 mM KCl and 10 mM MgCl2. The DNA and H-NS structure factors are obtained from the scattered intensities of samples having different solvent contrasts. Fig. 1 shows the intensities for H-NS–DNA in the two buffer systems at four different H2O/D2O solvent compositions. For all but the protein-matched samples, the scattering is dominated by protein. The structure factors are obtained from a fit to the complete data set consisting of the intensities pertaining to the four different solvent compositions. Notice that the contrasts of H-NS and DNA are matched in solvents of 43% and 64% D2O, respectively. Accordingly, the DNA and H-NS structure factors can also be obtained directly from the H-NS and DNA-matched samples, respectively, without fitting procedure. We found that the fitted and directly measured structure factors are identical within statistical accuracy. There is hence no uncertainty associated with obtaining the structure factors from a combination of intensities from different samples.The DNA structure factor multiplied by momentum transfer qSn is shown in panel A and B of Fig. 2 for the two buffer systems, respectively. In the plot of qSnversus q, for an infinitesimally thin cylinder a high q plateau is expected. However, no plateau is observed. The absence of q−1 scaling occurs as a result of the finite cross section of the DNA molecule. For the sample in T-buffer with 100 mM KCl, the DNA structure factor agrees with the high q limiting form of the rigid rod form factor with a radius of 0.8 nm.22 The sample is sufficiently diluted for absence of intermolecular interference in the present range of momentum transfer. In the presence of 10 mM MgCl2, the agreement between the DNA structure factor and the rod form factor is not as good. A distinct upturn in the DNA structure factor is observed at low values of momentum transfer q < 0.8 nm−1 (distances π/q > 4 nm). We attribute this upturn to H-NS mediated aggregation (bridging), which is known to be triggered by divalent cations.12,13 Besides experimental difficulties associated with the relatively weak scattering of DNA, it is unlikely that H-NS mediated bridging (and, hence, the upturn) can be eliminated by performing experiments at lower concentrations of DNA. For larger values of momentum transfer (q > 0.8 nm−1), the effect of aggregation disappears. We refrain from further interpretation of the DNA structure factor and focus on the structure of the nucleoprotein complex as revealed by the structure factor pertaining to H-NS.
The protein structure factor Sa is shown in panel A and B of Fig. 3 for H-NS–DNA in the absence and presence of magnesium ions, respectively. The H-NS to bp ratio follows from the normalisation of Sa and agrees with the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 set by the respective concentrations of DNA and H-NS. In the double logarithmic representation, Sa shows a characteristic shoulder at higher values of momentum transfer. This shoulder becomes less prominent, but does not disappear, in the presence of MgCl2. A similar shoulder, albeit at smaller q-values, was previously reported for another bacterial protein Hfq.7 This feature in the protein structure factor can be attributed to shell-like ordering of protein about DNA. As in the case of the DNA structure factor, Sa shows a small, but distinct low q upturn in the presence of MgCl2. The effect of divalent cations induced aggregation on the structure factors disappears however for q > 0.8 nm−1. Accordingly, the characteristic shoulder exhibited by Sa at larger values of q is not related to aggregation nor inter-complex solution structure. Information on the structure of the thus formed protein coat can be obtained by comparing coarse-grained model calculations with the low resolution experimental data. For this purpose, the protein structure factor is compared to the relevant form factor Sa = πaa2(q)/qL, with aa(q) being the Hankel transform of the radial amino acid density profile (see Materials and methods). In the model calculations, the predicted structure factors are convoluted with the instrumental resolution function.
6 set by the respective concentrations of DNA and H-NS. In the double logarithmic representation, Sa shows a characteristic shoulder at higher values of momentum transfer. This shoulder becomes less prominent, but does not disappear, in the presence of MgCl2. A similar shoulder, albeit at smaller q-values, was previously reported for another bacterial protein Hfq.7 This feature in the protein structure factor can be attributed to shell-like ordering of protein about DNA. As in the case of the DNA structure factor, Sa shows a small, but distinct low q upturn in the presence of MgCl2. The effect of divalent cations induced aggregation on the structure factors disappears however for q > 0.8 nm−1. Accordingly, the characteristic shoulder exhibited by Sa at larger values of q is not related to aggregation nor inter-complex solution structure. Information on the structure of the thus formed protein coat can be obtained by comparing coarse-grained model calculations with the low resolution experimental data. For this purpose, the protein structure factor is compared to the relevant form factor Sa = πaa2(q)/qL, with aa(q) being the Hankel transform of the radial amino acid density profile (see Materials and methods). In the model calculations, the predicted structure factors are convoluted with the instrumental resolution function.
Based on 150 bp DNA fragments and an H-NS to bp ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6, the intensity of free H-NS without long range order is a factor of 25 lower than the one of H-NS bound to DNA. Accordingly, free H-NS does not significantly contribute to the scattering at small angles. For the amino acid distribution in the radial direction away from the axis of the nucleoprotein complex, we have used three different models. In the first model, a Gaussian profile ρa(r) = exp(−r2/ra2)/(πra2) with cross-sectional radius of gyration ra [Hankel transform aa(q) = exp(−ra2q2/4)] is assumed. The corresponding model calculations are shown in the dashed curves in Fig. 3 (the radial profiles are shown in the insets). The use of a Gaussian profile results in a reasonable fit in the lower range of momentum transfer, but fails to predict the characteristic shoulder observed at higher values of q. The fit is not significantly affected by the (aggregation related) low q upturn in the presence of MgCl2. For the Gaussian model, we have optimised the value of ra. The results are shown in Table 3, together with the corresponding result reported for Hfq.7 The cross-sectional radius of gyration of the H-NS–DNA complex is significantly smaller than that for the Hfq hexamer, which can be attributed to the smaller molecular weight of the H-NS dimer. Furthermore, the cross-sectional dimension decreases by 35% with the addition of 10 mM MgCl2, which indicates a major structural rearrangement of the nucleoprotein complex.
6, the intensity of free H-NS without long range order is a factor of 25 lower than the one of H-NS bound to DNA. Accordingly, free H-NS does not significantly contribute to the scattering at small angles. For the amino acid distribution in the radial direction away from the axis of the nucleoprotein complex, we have used three different models. In the first model, a Gaussian profile ρa(r) = exp(−r2/ra2)/(πra2) with cross-sectional radius of gyration ra [Hankel transform aa(q) = exp(−ra2q2/4)] is assumed. The corresponding model calculations are shown in the dashed curves in Fig. 3 (the radial profiles are shown in the insets). The use of a Gaussian profile results in a reasonable fit in the lower range of momentum transfer, but fails to predict the characteristic shoulder observed at higher values of q. The fit is not significantly affected by the (aggregation related) low q upturn in the presence of MgCl2. For the Gaussian model, we have optimised the value of ra. The results are shown in Table 3, together with the corresponding result reported for Hfq.7 The cross-sectional radius of gyration of the H-NS–DNA complex is significantly smaller than that for the Hfq hexamer, which can be attributed to the smaller molecular weight of the H-NS dimer. Furthermore, the cross-sectional dimension decreases by 35% with the addition of 10 mM MgCl2, which indicates a major structural rearrangement of the nucleoprotein complex.
| r a (nm) | r 2 (nm) | r b (nm) | F | |
|---|---|---|---|---|
| H-NS/KCl | 2.3 ± 0.2 | 3.2 ± 0.2 | 0.52 ± 0.02 | 0.13 ± 0.02 |
| H-NS/KCl/MgCl2 | 1.5 ± 0.1 | 2.5 ± 0.2 | 0.90 ± 0.05 | 0.48 ± 0.05 |
| Hfq/KCl | 5.0 ± 0.5 | 6.0 ± 0.5 | — | — |
A deficiency of the Gaussian model is that it ignores the depletion of protein density at the core of the complex. In the second model, H-NS is assumed to form a cylindrical coat surrounding the duplex, with inner and outer radii r1 and r2, respectively. The radial amino acid distribution is, hence, constant for r1 < r < r2, and given by ρ(r)π(r22 − r12) = 1. As was previously reported, the coat model reproduces the structure factor of Hfq well.7 For H-NS, the shell-like radial profiles and corresponding model calculations are shown by the dotted curves in Fig. 3. In the model calculations, the inner radius of the protein distribution was set to the outer radius of the DNA molecule, that is r1 = 1.0 nm. The outer radius of the complex r2 was optimised to reproduce the position of the shoulder in q-space (results are shown in Table 3). In the presence of 10 mM MgCl2, the shoulder is shifted towards higher values of momentum transfer due to a decrease in outer radius. Although its position in q-space can be well reproduced, the coat model does not provide a good description of the intensity and shape of the shoulder. This fit cannot be improved by a decrease in the value of r1 (for instance, to account for insertion of a short C-terminal loop into the minor groove, see below) and/or by a helical distribution of H-NS around the DNA molecule.
The poor fit of the coat model and the reduction in cross-sectional radius of gyration with the addition of magnesium salt suggest that a fraction of H-NS is distributed at a smaller distance, closer to the axis of the complex than the DNA outer radius of 1.0 nm. The major groove of B-form DNA has a depth and width of 0.9 and 1.2 nm, respectively, and can accommodate (part of) a small protein such as H-NS.23 In our third model, the amino acid density follows a bimodal radial distribution. Sections of the DNA molecule are surrounded by protein, as in the second model. For this coat of protein, the inner radius is set to r1 = 1.0 nm and the outer radius r2 is obtained from the fit of the position of the shoulder (Table 3). In other sections of the DNA molecule, H-NS is bound to the major groove. The corresponding amino acid profile is assumed to be step-like with an arbitrary width of 0.5 nm and centred at a distance rb from the axis of the complex. We have verified that the structure factor is most sensitive to fraction F and binding distance rb of major groove bound H-NS, whereas the width of the profile is of lesser importance. Long range correlation and, hence, interference between sections of coat-forming and major groove bound H-NS are neglected. The radial profiles and corresponding model calculations with optimised values of F and rb (Table 3) are shown by the solid curves in Fig. 3. The experimental structure factors and, in particular, the effect of MgCl2, are well reproduced. In T-buffer with 100 mM KCl, a small fraction of H-NS is major groove bound and distributed mid-way between the central axis and outer radius of the DNA molecule. However, most of H-NS covers the region next to the duplex. With the addition of 10 mM MgCl2, the complex tightens as shown by a reduction in outer diameter. Furthermore, about half of H-NS is now major groove bound and distributed close to the phosphates of the DNA molecule.
4 Discussion
Our most important result is the determination of the contribution to the scattering from H-NS. This corresponds to a cylindrical nucleoprotein complex with an H-NS to DNA base-pair ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6. We have evaluated and tested various amino acid density profiles in the radial direction away from the axis of the complex. A cross-sectional radius of gyration is obtained from a fit of the Gaussian model in the low range of momentum transfer. The Gaussian model fails however to predict the characteristic shoulder observed in the protein structure factor at higher values of momentum transfer. This shoulder is related to the cut-off in amino acid density at the outer radius of the complex.7 A radial distribution representing a single coat of H-NS surrounding the duplex does however not provide a satisfactory fit. Fairly good agreement between coarse-grained model calculations based on the radial density profiles and experimental protein structure factors is obtained using a heterogeneous, two-state binding model with sections of the duplex surrounded by protein and other sections with protein bound to the major groove. In a buffer of monovalent salts, most of H-NS covers the region next to the duplex. With the addition of magnesium chloride, we observe a structural rearrangement through a decrease in diameter and increase in the fraction of major groove bound H-NS.
6. We have evaluated and tested various amino acid density profiles in the radial direction away from the axis of the complex. A cross-sectional radius of gyration is obtained from a fit of the Gaussian model in the low range of momentum transfer. The Gaussian model fails however to predict the characteristic shoulder observed in the protein structure factor at higher values of momentum transfer. This shoulder is related to the cut-off in amino acid density at the outer radius of the complex.7 A radial distribution representing a single coat of H-NS surrounding the duplex does however not provide a satisfactory fit. Fairly good agreement between coarse-grained model calculations based on the radial density profiles and experimental protein structure factors is obtained using a heterogeneous, two-state binding model with sections of the duplex surrounded by protein and other sections with protein bound to the major groove. In a buffer of monovalent salts, most of H-NS covers the region next to the duplex. With the addition of magnesium chloride, we observe a structural rearrangement through a decrease in diameter and increase in the fraction of major groove bound H-NS.
The scattering data adds quantitative information to existing knowledge of the H-NS–DNA complex and, in particular, the effect of divalent cations. The 83 N-terminal region is connected through a flexible linker to a short C-terminal DNA binding region (residues 91–137).26,27 Furthermore, the N-terminal region contains a primary dimerization site 1 (residues 1–46) and secondary oligomerization site 2 (residues 67–83). Head-to-head dimers are formed by interaction of sites 1. Dimeric H-NS binds selectively to the minor groove by insertion of the C-terminal loop.19 The head-to-head dimerization through sites 1 together with tail-to-tail oligomerization of sites 2 creates a chain of linked H-NS molecules that form a superhelical scaffold.24 The resulting nucleoprotein filament has been proposed to be critical for the gene silencing function.14–17 It should also be noted that the linked protein coat significantly stiffens the filament, as shown by an increase in bending persistence length from around 60 to 130 nm.10–13 A model of H-NS oligomerization and the filament are shown in Fig. 4. In a buffer of monovalent salts, we observed that most of the DNA molecules or sections thereof are surrounded by H-NS. This coat of protein covers the area next to the duplex, which agrees with selective binding of dimeric H-NS with the C-terminal loop to the minor groove and oligomerization of the N-terminal region to form a filament. The outer diameter of 6.4 ± 0.4 nm shows that the complex is relatively slender. Furthermore, the contour length of the complex is close to that of naked DNA, which shows that the superhelical protein scaffold is deformed with respect to the structure observed for mutants lacking the C-terminal DNA-binding region.24,28 A small fraction of H-NS is bound to the major groove. The latter fraction has likely a different biological function, as indicated by the effect of divalent salts on the DNA-compaction properties of H-NS.
 | ||
| Fig. 4 (A) An oligomer of three 83 N-terminal regions of H-NS from S. typhymurium, connected through dimerization interface site 1 and oligomerization interface site 2 (PDB 3NR7).24 (B) C-terminal DNA binding region from E. coli Ler (PDB 2LEV).25 (C) Model of H-NS bound to DNA through its C-terminal loop and linked through site 1 and site 2 interfaces. (D) Model of an inter-duplex bridge through site 2 interaction. The N-terminal region is folded into the major groove. (E) As in (D), but for a bridge through site 1 interaction. Only the central part of the N-terminal region is folded into the major groove. | ||
With the addition of magnesium chloride, H-NS-mediated bridges between distal DNA segments are formed. Bridging has been proposed to occur by interfacing sites 1 or, more recently, sites 2.24,29 Furthermore, the persistence length of the complex decreases and takes a value of around the one pertaining to bare DNA.10–12 Bridging results in compaction and, eventually, a collapse of the DNA molecule into a condensed state.13 Here, we observe a concurrent change in the structure of the nucleoprotein complex, that is the complex tightens and the fraction of H-NS bound to the major groove increases. The decrease in persistence length indicates that the chain of linked H-NS proteins is broken by disruption of either interfacial site 1 or 2. If the chain is broken at interfacial site 1, the N-terminal region may be folded into the major groove, facilitated by the flexible linker, and stabilised by hydrophobic interaction with the stacked base pairs. A small number of broken links explains the small fraction of major groove bound H-NS even in the absence of divalent salts. However, as shown by the DNA compaction properties, major groove bound H-NS is involved in bridging rather than the formation of a filament. A major groove bound H-NS monomer can form a bridging link through site 2 with a monomer bound to another DNA segment, resulting in short range inter-DNA segment attraction (a model is shown in Fig. 4D). Alternatively, the chain of linked H-NS proteins can be broken at site 2 and the bridge can be formed through interfacing sites 1 (Fig. 4E). The latter scenario is less likely, because a larger section of the protein (site 1, residues 1–46) needs to be accommodated next to the duplex to form the bridging link at the cost of reduced stabilising hydrophobic interaction with the stacked base pairs. Both models explain major groove binding, tightening of the complex, increase in bending flexibility, and propensity for bridging. Unfortunately, the exact structure of the H-NS mediated bridges cannot be determined based on the present low resolution diffraction data.
The two binding states identified here may correspond to two distinct biological functions. Coat-forming H-NS may be involved in gene silencing through the formation of a filament, whereas major groove bound H-NS might have a predominant function in genome organisation through protein-mediated DNA segment interaction.
Acknowledgements
This work is supported by the Singapore Ministry of Education grant MOE2014-T2-1-001. The Institut Laue–Langevin is thanked for providing the neutron scattering facilities. We thank V. Dahirel for his contribution in cloning and purification of H-NS. J. Yan and S. Rimsky are thanked for discussions.References
- C. L. Woldringh and N. Nanninga, J. Struct. Biol., 2006, 156, 273–283 CrossRef CAS PubMed.
- J. R. C. van der Maarel, L. C. Groot, M. Mandel, W. Jesse, G. Jannink and V. Rodriguez, J. Phys. II, 1992, 2, 109–122 CrossRef CAS.
- L. C. A. Groot, M. E. Kuil, J. C. Leyte, J. R. C. van der Maarel, J.-P. Cotton and G. Jannink, J. Phys. Chem., 1994, 98, 10167–10172 CrossRef CAS.
- K. Kassapidou, W. Jesse, M. E. Kuil, A. Lapp, S. Egelhaaf and J. R. C. van der Maarel, Macromolecules, 1997, 30, 2671–2684 CrossRef CAS.
- S. S. Zakharova, S. U. Egelhaaf, L. B. Bhuiyan, C. W. Outhwaite, D. Bratko and J. R. C. van der Maarel, J. Chem. Phys., 1999, 111, 10706–10716 CrossRef CAS.
- L. Dai, Y. Mu, L. Nordenskiöld, A. Lapp and J. R. C. van der Maarel, Biophys. J., 2007, 92, 947–958 CrossRef CAS PubMed.
- K. Jiang, C. Zhang, D. Guttula, F. Liu, J. A. van Kan, C. Lavelle, K. Kubiak, A. Malabirade, A. Lapp, V. Arluison and J. R. C. van der Maarel, Nucleic Acids Res., 2015, 43, 4332–4341 CrossRef CAS PubMed.
- H. Wang, J. C. Ayala, J. A. Benitez and A. J. Silva, PLoS One, 2015, 10, e0118295 Search PubMed.
- C. Kahramanoglou, A. S. N. Seshasayee, A. I. Prieto, D. Ibberson, S. Schmidt, J. Zimmermann, V. Benes, G. M. Fraser and N. M. Luscombe, Nucleic Acids Res., 2011, 39, 2073–2091 CrossRef CAS PubMed.
- R. T. Dame, C. Wyman and N. Goosen, Nucleic Acids Res., 2000, 28, 3504–3510 CrossRef CAS PubMed.
- R. Amit, A. B. Oppenheim and J. Stavans, Biophys. J., 2003, 84, 2467–2473 CrossRef CAS PubMed.
- Y. Liu, H. Chen, L. J. Kenney and J. Yan, Genes Dev., 2010, 24, 339–344 CrossRef CAS PubMed.
- C. Zhang, D. Guttula, F. Lui, P. P. Malar, S. Y. Ng, L. Dai, P. Doyle, J. A. van Kan and J. R. C. van der Maarel, Soft Matter, 2013, 9, 9593–9601 RSC.
- C. Badaut, R. Williams, V. Arluison, E. Bouffartigues, B. Robert, H. Buc and S. Rimsky, J. Biol. Chem., 2002, 277, 41657–41666 CrossRef CAS PubMed.
- S. Stella, R. Spurio, M. Falconi, C. L. Pon and C. O. Gualerzi, EMBO J., 2005, 24, 2896–2905 CrossRef CAS PubMed.
- E. Bouffartigues, M. Buckle, C. Badaut, A. Travers and S. Rimsky, Nat. Struct. Mol. Biol., 2007, 14, 441–448 CAS.
- C. J. Lim, S. Y. Lee, L. J. Kenney and J. Yan, Sci. Rep., 2012, 2, 509 Search PubMed.
- D. Tippner and R. Wagner, J. Biol. Chem., 1995, 270, 22243–22247 CrossRef CAS PubMed.
- B. R. G. Gordon, Y. Li, A. Cote, M. T. Weirauch, P. Ding, T. R. Hughes, W. W. Navarre, B. Xia and J. Liu, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 10690–10695 CrossRef CAS PubMed.
- L. Wang, M. Ferrari and V. A. Bloomfield, Biotechniques, 1990, 9, 24–27 CAS.
- B. Jacrot, Rep. Prog. Phys., 1976, 39, 911–953 CrossRef CAS.
- J. R. C. van der Maarel and K. Kassapidou, Macromolecules, 1998, 31, 5734–5739 CrossRef CAS.
- V. Bloomfield, D. Crothers and I. Tinoco, Nucleic Acids; Structure, Properties and Functions, University Science Books, Sausilito, 2000 Search PubMed.
- S. T. Arold, P. G. Leonard, G. N. Parkinson and J. E. Ladbury, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 15728–15732 CrossRef CAS PubMed.
- T. N. Cordeiro, H. Schmidt, C. Madrid, A. Juarez, P. Bernado, C. Griesinger, J. Garcia and M. Pons, PLoS Pathog., 2011, 7, e1002380 CAS.
- C. J. Dorman, Nat. Rev. Microbiol., 2004, 2, 391–400 CrossRef CAS PubMed.
- M. Sette, R. Spurio, E. Trotta, C. Brandizi, A. Brandi, C. L. Pon, G. Barbato, R. Boelens and C. O. Gualerzi, J. Biol. Chem., 2009, 284, 30453–30462 CrossRef CAS PubMed.
- R. S. Winardhi, R. Gulvady, J. L. Mellies and J. Yan, J. Biol. Chem., 2014, 289, 13739–13750 CrossRef CAS PubMed.
- R. T. Dame, M. C. Noom and G. J. L. Wuite, Nature, 2006, 444, 387–390 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5sm03076e |
| This journal is © The Royal Society of Chemistry 2016 |