 Open Access Article
Open Access ArticleMechanical signatures of microbial biofilms in micropillar-embedded growth chambers†
S. C.
Chew
ab,
B.
Kundukad
c,
W. K.
Teh
a,
P.
Doyle
cd,
L.
Yang
ae,
S. A.
Rice
aef and
S.
Kjelleberg
*aef
aSingapore Centre for Environmental Life Sciences Engineering (SCELSE), Nanyang Technological University, Singapore. E-mail: LASKJELLEBERG@ntu.edu.sg
bInterdisciplinary Graduate School, Nanyang Technological University, Singapore
cBioSystems and Micromechanics IRG, Singapore-MIT Alliance for Research and Technology, National University of Singapore, Singapore
dDepartment of Chemical Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA
eSchool of Biological Sciences, Nanyang Technological University, Singapore
fCentre for Marine Bio-Innovation and School of Biotechnology and Biomolecular Sciences, University of New South Wales, Australia
First published on 3rd May 2016
Abstract
Biofilms are surface-attached communities of microorganisms embedded in an extracellular matrix and are essential for the cycling of organic matter in natural and engineered environments. They are also the leading cause of many infections, for example, those associated with chronic wounds and implanted medical devices. The extracellular matrix is a key biofilm component that determines its architecture and defines its physical properties. Herein, we used growth chambers embedded with micropillars to study the net mechanical forces (differential pressure) exerted during biofilm formation in situ. Pressure from the biofilm is transferred to the micropillars via the extracellular matrix, and reduction of major matrix components decreases the magnitude of micropillar deflections. The spatial arrangement of micropillar deflections caused by pressure differences in the different biofilm strains may potentially be used as mechanical signatures for biofilm characterization. Hence, we submit that micropillar-embedded growth chambers provide insights into the mechanical properties and dynamics of the biofilm and its matrix.
Introduction
Microorganisms excrete and embed themselves in a matrix of extracellular polymeric substances (EPS)1 to form surface-attached communities, or biofilms, as their predominant lifestyle in nature.2 Biofilms play essential roles in sustaining natural environments and human society, including the remediation of natural habitats, treating wastewater and bioleaching.3 Biofilms can also be problematic in industry; for example, biomass accumulation in pipes disrupts flow and leads to corrosion, contamination of production lines, compromises food quality, and biofilm formation on reverse osmosis membranes limits the production of clean water and increases running costs.4 Biofilms have a significant impact on healthcare, where the US National Institutes of Health (NIH) has estimated that more than 80% of chronic infections and 65% of microbial infections are biofilm-associated (Program Announcement Number PA-03-047, National Institutes of Health).Biofilm formation is a highly regulated process, whereby microorganisms employ physiological cooperation and spatial organization to increase both their metabolic efficiency and adaptation to changes in their local environment.5 The result of this process is a multicellular structure that is usually heterogeneous in architecture. Cell growth and death, EPS production and degradation are factors that generate mechanical forces that move biomass to expand or remodel the biofilm. However, research into measuring mechanical forces generated by the biofilm, specifically how they contribute to the development of the biofilm structure, is still in its infancy for many biofilms. A recent study has found that Bacillus subtilis floating pellicles maintain a low internal stress that drives biofilm spreading after relaxation from confinement and recovery during biofilm ablation.6 For B. subtilis grown at the air–solid surface interface, such as on agar media, biofilm spreading is mediated by the osmotic pressure resulting from the EPS absorbing water, rather than the internal pressure caused by colony growth.7 Lateral mechanical forces are spatially focused by localized cell death in B. subtilis, driving macroscopic movement and vertical buckling of the biofilm matrix. This results in a wrinkled morphology that may increase resistance to liquid wetting and gas penetration.8,9
Moreover, the mechanical strength and viscoelasticity of the EPS is expected to have an impact on external and internal mechanical forces experienced by the biofilm, and thus influence any movement or rearrangement of biomass and affect biofilm structure and morphology. For example, the EPS of P. aeruginosa biofilms can shear under flow, creating string-like extensions that eventually result in the development of streamers.10Pseudomonas aeruginosa biofilms are also enhanced in surface spreading and streamer formation when loose viscoelastic matrices are produced, and are reduced in surface spreading and streamer formation when densely crosslinked elastic matrices are produced.11 As such, it is important to study the mechanical forces or pressure that drive the spreading and self-organization of these microbial populations, which can also provide a conceptual framework for other microbial systems.
Micropillar arrays were developed for the study and measurement of cellular/subcellular traction forces that eukaryotic cells exert upon adhesion to substrates. In the standard micropillar setup and model (Fig. 1a), the cell is required to adhere to, and therefore leverage on, more than one micropillar for attachment and traction. The micropillar array has been extensively used for studies of cell spreading and single cell migration, as well as movement of monolayers of mammalian cells. For eukaryotic cells, the cellular/subcellular traction forces are generated by the cytoskeletal motor protein myosin II, which causes re-arrangement of actin filaments, when coupled to adhesion sites of the cell.12,13 In contrast, apart from the measurement of cooperative retraction forces by the type IV pili in Neisseria gonorrhoeae, for which large forces of 200 pN to 1 nN have been recorded,14 micropillars have not been used in force studies of microbial cells. Unlike mammalian cells, microbial cells are considered rigid, with the Young's modulus (E) of live Escherichia coli being recorded as 1.9 ± 0.9 to 3.0 ± 0.6 MPa, and 6.1 ± 1.5 MPa for dead E. coli.15 As such, they have well-defined shapes that do not deform easily and are unlikely to generate high traction forces within the cell body. In addition, micropillar dimensions are usually in the order of 1–3 μm in diameter, and require sufficient spacing intervals for deflections to be measured. Thus, most microbial cells are too small to attach to and interact with the multiple micropillars required to generate force measurements. However, within the biofilm, cells are collectively held together by an extracellular matrix, of which intercellular forces between cells have been reported to range from 6.5–6.8 nN, based on atomic force microscopy.16 The Young's modulus (E) for biofilms varies widely, with reported values of 37.82 ± 5.87 kPa17 and 25.0 ± 2.5 kPa8 for early stage laboratory biofilms grown on agar, and 200 to 9000 kPa18 for environmental biofilms.
The multicellular biofilm can spread over a large area and grow to hundreds of micrometers in thickness. The extracellular matrix secreted by the cells within the biofilm is expected to be the major component that binds to multiple micropillars while holding the cells together (Fig. 1b). We hypothesize that once the biofilm is formed, forces generated by multicellular microbial behaviour involving cell growth, death, motility and differentiation would create internal mechanical forces or pressure. While a uniform pressure generates equal lateral forces in all directions and does not deflect the micropillars, the heterogeneous growth and development of the biofilm may produce net mechanical forces or differential pressure, sufficient to deflect the micropillars.
This work describes the application of micropillar-embedded growth chambers as a tool for the mechanical characterization of bacterial biofilms. We investigated biofilms formed by several medical and environmental model bacterial species, and mutants of these, defective in the production of key matrix components, including E. coli strains causing urinary tract infections,19 as well as non-virulent strains used in the industrial production of recombinant therapeutics. P. aeruginosa is a common opportunistic pathogen. The mucoid type, in which the bacterium overexpresses alginate along with exopolysaccharides Pel and Psl in its matrix, is often isolated from cystic fibrosis patients where it is a major biofilm forming bacterial species.20Staphylococcus aureus and Staphylococcus epidermidis often cause biofilm-associated wound infections and device-related infections.21,22 The environmental bacterium Shewanella oneidensis reduces heavy metals and many complex xenobiotics.23 Herein, we show that micropillars can measure forces at the substratum related to biofilm growth and matrix mechanical properties. The various biofilms generate deflection patterns in the micropillar array that are reflective of their growth dynamics and EPS mechanical properties.
Experimental
Biofilm cultivation
The bacterial strains used in this study include E. coli SAR18 F+; P. aeruginosa PAO1ΔmucA, PAO1ΔmucAΔpslBCD and PAO1ΔmucAΔpelA; S. aureus 15981; S. epidermidis 1457 and 1457ΔatlE; S. oneidensis MR-1 (Table 1). Overnight cultures of bacterial strains were grown at their optimal temperatures and growth medium to an OD600 = 2.0. For E. coli SAR18 F+, P. aeruginosa ΔmucA, PAO1 ΔmucAΔpslBCD and PAO1 ΔmucAΔpelA, growth was at 37 °C in Luria Broth (LB) medium. For S. aureus 15981, S. epidermidis 1457 and S. epidermidis 1457ΔatlE, growth was at 37 °C in Tryptic Soy Broth (TSB) medium. S. oneidensis MR-1 was grown at 30 °C in LB medium. Subsequently, for biofilm formation, 10 μL of overnight cultures were added to the wells of the growth chambers with 190 μL of medium to a final OD600 = 0.1, and incubated for 24 h under static conditions at the optimal temperature of 37 °C or 30 °C for each organism as described above. The growth curves of the bacterial strains using the above cultivation conditions are provided in Fig. S1 (ESI†).| Strain | Description | Ref. |
|---|---|---|
| SAR18 F+ | E. coli CSH26 wild-type, carrying transfer constitutive IncF plasmids that promote cell–cell adhesion and biofilm formation mediated by F pili | 24 |
| PAO1ΔmucA | Mucoid P. aeruginosa PAO1, expressing alginate, Pel and Psl exopolysaccharides | 25 |
| PAO1ΔmucAΔpelA | Psl mutant of mucoid PAO1, expressing alginate and Pel exopolysaccharides only | 26 |
| PAO1ΔmucAΔpslBCD | Pel mutant of mucoid PAO1, expressing alginate and Psl exopolysaccharides only | 26 |
| 15981 | S. aureus wild-type | 26 |
| 1457 | S. epidermidis wild-type | 27 |
| 1457ΔatlE | Autolysin (atlE) mutant of S. epidermidis 1457, unable to release extracellular DNA (eDNA) into matrix | 27 |
| MR-1 | S. oneidensis wild-type | 23 |
Micropillar arrays
PDMS (polydimethylsiloxane) micropillar arrays were moulded from silicon masters made with conventional high-resolution photolithography and deep reactive ion-etching techniques.28 The micropillars had a height (H) of 3 μm, diameter (D) of 1.5 μm and center-to-center spacing of 4 μm. The Young's modulus (E) of the micropillars was 1.0 ± 0.3 MPa, as verified using atomic force microscopy. The PDMS micropillar arrays were established at the bottom of growth chambers to form 300 μL wells, 9.4 mm in width and 10.7 mm in length. The growth chambers were placed in 100% ethanol and sonicated to ensure the micropillars remained in upright positions, and to avoid collapse of the micropillars during introduction of high surface tension fluids. The ethanol solution was replaced with sterile water, and then LB or TSB medium to a final volume of 190 μL in each well.Microscopy and imaging
For the imaging of micropillar deflections, bright-field Z-stack micrographs of the micropillars after allowing for 24 h of bacterial attachment and biofilm growth were captured (ZEISS Axio Imager M1). Biofilms were stained with Congo Red (Sigma-Aldrich, Singapore) to improve the contrast between the biofilm and micropillars. The positions of the micropillar tops and bottoms were located by their centroid positions using a Gaussian or Mexican Hat filter (background subtraction) in IMARIS software (Bitplane, Zürich). The deflection vector r is given by the difference in positions: | (1) |
For the biomass quantification of biofilms formed by the respective strains after 24 h of growth, confocal images of the biofilm were captured (Zeiss LSM780 confocal scanning laser microscope) and analyzed by COMSTAT (http://www.comstat.dk).29,30 Biofilms were cultivated in chambers without micropillars. The Gram-negative E. coli, P. aeruginosa and S. oneidensis were tagged with the green fluorescent protein (Gfp) for visualization and quantification of live biomass, and stained with propidium iodide (PI) for visualization and quantification of dead biomass. The Gram-positive S. aureus and S. epidermidis were stained with SYTO9 and PI for visualization and quantification of live and dead biomass.
Force calculations
In the case where the mammalian cell body is located on top of the micropillars, the micropillar is modelled as a cantilevered beam deflected by a horizontal traction force applied on the micropillar top, f (see Fig. 1a). For small deformations, the force required to deflect the micropillar maybe approximated by Hooke's law, using the Euler–Bernoulli beam theory and the following equation:31 | (2) |
 | (3) |
 | (4) |
 | (5) |
 | (6) |
 | (7) |
 | (8) |
Results and discussion
Distribution of pressure differences in various biofilms
The bacterial strains attached to the micropillars at the substratum of the growth chambers and formed biofilms that deflected the micropillars over 24 h. The deflection magnitudes of 120 micropillars within a 50 × 50 μm area of substratum from at least three different experimental replicates for each strain, were used to calculate the differential pressure applied to the micropillar over 24 h. A histogram was compiled to compare the distribution of pressure differences (Fig. 2), and the average, maximum and standard deviation of differential pressure were calculated for biofilms of each wild-type and mutant strain (Table 2). For mucoid P. aeruginosa (PAO1ΔmucA) and S. epidermidis, most of the differences in pressure after 24 h of biofilm growth fell within the 4–8 kPa range. This range accounted for 54.8 ± 9.2% and 39.0 ± 7.4%, of the pressure differences in P. aeruginosa and S. epidermidis, respectively. For E. coli and S. aureus, most of the pressure differences fell within the 8–12 kPa range, accounting for 46.4 ± 5.4% and 39.6 ± 3.8%, respectively. For S. oneidensis, a majority of 95 ± 6% of pressure differences were below 4 kPa. E. coli had the highest average differential pressure at 8.5 ± 0.1 kPa. This was then followed by S. aureus at 8.0 ± 0.6 kPa, S. epidermidis at 7.9 ± 0.6 kPa, P. aeruginosa at 7.2 ± 0.2 kPa and S. oneidensis at 1.8 ± 0.6 kPa. The average differential pressure may be reflective of the heterogeneity of the particular species within the biofilm. In this study E. coli, S. aureus and S. epidermidis formed a heterogeneous biofilm with many microcolonies and channels. Mucoid P. aeruginosa also formed heterogeneous biofilms, but with smaller microcolonies and more undifferentiated areas. S. oneidensis did not form differentiated biofilms under our growth conditions.| Strain | Differential pressurea (kPa) | ||
|---|---|---|---|
| Average | Maximum | S.D.b | |
| a Calculated from 120 micropillars within a 50 × 50 μm area. At least three biological replicates were used. b Standard deviation. | |||
| E. coli SAR18 F+ | 8.5 ± 0.1 | 16.1 ± 2.7 | 3.1 |
| P. aeruginosa PAO1ΔmucA | 7.2 ± 0.2 | 12.0 ± 1.0 | 2.3 |
| P. aeruginosa PAO1ΔmucAΔpelA | 3.1 ± 0.7 | 6.5 ± 1.0 | 1.2 |
| P. aeruginosa PAO1ΔmucAΔpslBCD | 6.2 ± 0.3 | 10.3 ± 1.5 | 1.8 |
| S. aureus 15981 | 8.0 ± 0.6 | 18.7 ± 2.2 | 3.2 |
| S. epidermidis 1457 | 7.9 ± 0.6 | 20.0 ± 3.1 | 3.7 |
| S. epidermidis 1457ΔatlE | 5.3 ± 0.9 | 20.5 ± 5.0 | 3.7 |
| S. oneidensis MR-1 | 1.8 ± 0.6 | 5.3 ± 1.6 | 1.0 |
While E. coli had the highest average differential pressure, S. aureus and S. epidermidis were able to achieve differential pressures greater than 20 kPa. However, this accounted for less than 1% of the differential pressure. S. epidermidis had the highest maximum differential pressure and standard deviation, followed by S. aureus, E. coli, P. aeruginosa and S. oneidensis (Table 2).
Differential pressure arises from the pressure of a growing biofilm biomass exerting itself onto the micropillars. To examine the correlation between differential pressure and the total biofilm biomass generated by the strains, we measured the biomass of static biofilm cultures for each strain (Fig. 3). The live and dead measurements were combined to determine the total biomass. Among wild-type strains and mucoid P. aeruginosa, S. aureus and S. epidermidis biofilms had the highest total biomass with 5.8 ± 0.5 μm3 μm−2 and 5.4 ± 0.1 μm3 μm−2, respectively. E. coli displayed the second highest total biomass at 5.1 ± 1 μm3 μm−2. Biofilms of PAO1ΔmucA, S. aureus and S. epidermidis had significant proportions of dead biomass at 62 ± 7%, 49 ± 2% and 43 ± 8% respectively. E. coli biofilms displayed a small fraction, 10 ± 5%, of dead biomass. This suggests that cell death was not a major factor for its higher average differential pressure, as would be predicted based on previous data for B. subtilis.8 PAO1ΔmucA and S. oneidensis generated the second lowest and lowest total biomass at 3.0 ± 0.1 μm3 μm−2 and 1.7 ± 0.3 μm3 μm−2, respectively, and may explain their comparatively lower average differential pressures. These results indicate that although the generation of total biomass could partly account for the differential pressure, other species-specific biofilm matrix factors might also be involved.
Matrix components of the biofilm strongly influence differential pressure generated by the biofilm
EPS mechanical properties were hypothesized to influence the deflection of the micropillars. To examine whether micropillars can be used to investigate the impact of specific EPS components on differential pressure in the biofilm, mucoid P. aeruginosa, S. epidermidis strains, and their respective matrix mutant strains, were assessed (Table 1 and Fig. 4). P. aeruginosa ΔmucAΔpelA and P. aeruginosa ΔmucAΔpslBCD mutants are unable to synthesize the major P. aeruginosa matrix components Pel and Psl exopolysaccharides, respectively.26,34 Both S. aureus and S. epidermidis release a large amount of eDNA into their matrix, which contributes significantly to biofilm formation.27,35S. epidermidis ΔatlE mutants are unable to release eDNA into the biofilm matrix and hence are expected to produce weaker biofilms.27 | ||
| Fig. 4 Comparison of quartiles, maximum and minimum of differential pressure data for mucoid P. aeruginosa, wild-type S. epidermidis and their respective EPS mutant strains. | ||
The average and maximum differential pressure measured for the P. aeruginosa ΔmucAΔpelA biofilms were 3.1 ± 0.7 and 6.5 ± 1.0 kPa, respectively (Table 2 and Fig. 4). The average and maximum differential pressure measured for the P. aeruginosa ΔmucAΔpslBCD biofilms were 6.2 ± 0.3 and 10.3 ± 1.5 kPa, respectively (Table 2 and Fig. 4). Thus, the loss of the exopolysaccharide, Pel, significantly impaired the ability of the biofilm to deflect the micropillars, whereas loss of the exopolysaccharide, Psl, reduced the differential pressure to a lesser extent, compared to the P. aeruginosa ΔmucA parent strain. In the present study, the live and total biomasses of P. aeruginosa ΔmucAΔpslBCD, at 3.5 ± 0.6 μm3 μm−2 and 6.0 ± 0.1 μm3 μm−2, respectively, were much greater than for the P. aeruginosa ΔmucAΔpelA strain, at 0.8 ± 0.1 μm3 μm−2 and 3.9 ± 0.1 μm3 μm−2, respectively, and the P. aeruginosa ΔmucA strain, at 1.1 ± 0.2 μm3 μm−2 and 3.0 ± 0.1 μm3 μm−2, respectively (Fig. 3). This finding agrees with the report that the Pel polysaccharide is important for lateral growth and spreading,11 which would increase the overall pressure in the biofilm. In contrast, the Psl exopolysaccharide increased the cross-linking density and elasticity of the matrix, resulting in increased mechanical stiffness that restricted lateral growth and spreading. Biofilms containing the Pel polysaccharide were also shown to vary in rheology in different locations and over time, whereas biofilms expressing only Psl were spatially and temporally homogeneous in rheology.11 This may also explain the relatively larger contribution of Pel to differential pressure, compared to Psl. The expression of both Pel and Psl in P. aeruginosa ΔmucA biofilms had a synergistic effect on increasing pressure differences (Fig. 4), which may act to improve overall biofilm dynamics and formation.
eDNA is another well-known matrix component, crucial for maintaining the biofilm mechanical properties. The S. epidermidis eDNA deficient ΔatlE mutant biofilms showed a decreased ability to deflect the micropillars, with an average differential pressure of 5.2 ± 0.9 kPa, compared to 7.9 ± 0.6 kPa for the parent strain. In spite of this, a high differential pressure could still be achieved in the S. epidermidis ΔatlE biofilm. Indeed, the maximum differential pressure was 20.5 ± 5.0 kPa, which suggests that other major biofilm matrix components (e.g. polysaccharide intercellular adhesin) played a more important role in generating pressure differences.
Characteristic deflection patterns of bacterial strains are caused by biofilm heterogeneity and microcolony formation
Deflections of 120 micropillars within a 50 × 50 μm area were plotted as vectors (black arrows) on a 2D Cartesian plane for the mucoid P. aeruginosa and wild-type strains (Fig. 5). Micropillars in the control chamber remained undeflected and generated null vectors (Fig. 5a). E. coli SAR18 F+ (Fig. 5b), P. aeruginosa PAO1ΔmucA (Fig. 5c), S. aureus 15891 (Fig. 5d) and S. epidermidis 1457 (Fig. 5e) developed heterogeneous biofilms that exhibited extensive cell clustering and microcolony formation. The formation of such microcolonies deflected the micropillars in various directions and disrupted the regular micropillar positioning. As a measure of regularity in the deflections and to examine how similar in direction the vectors were across the different biofilms, the cosine similarity of each vector to its right horizontal and lower vertical neighbour was calculated according to the following equation: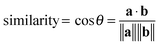 | (9) |
E. coli, mucoid P. aeruginosa, S. aureus and S. epidermidis 1457 biofilms displayed low average similarities of 0.12 ± 0.13, 0.11 ± 0.06, 0.17 ± 0.10 and 0.22 ± 0.07, respectively. The average angles between the neighbouring vectors were nearly orthogonal to each other at 83.2 ± 8.0°, 83.8 ± 3.8°, 80.0 ± 5.8° and 77.3 ± 4.0°, respectively. In contrast to the other species, S. oneidensis (Fig. 5f) formed a thin and homogenous biofilm without any microcolonies and as such, the distance between, and regularity of, the micropillars were largely maintained. On average, the deflection vectors showed a high similarity of 0.79 ± 0.05 and angle of 37.8 ± 5.1° with neighbouring vectors. Thus, the similarity between neighbouring vectors can be a measure of microcolony formation, with a high similarity indicating the presence of very few or no microcolonies.
A red contour, as approximated from a Z-stack of bright-field images, was drawn along the high cell density areas and microcolonies on the deflection vector map (Fig. 5). For E. coli and mucoid P. aeruginosa biofilms, micropillars were deflected within the microcolonies, as well as for the flat, undifferentiated areas of the biofilm. For S. aureus and S. epidermidis biofilms, the lengths of the deflection vectors varied widely along the contour of the biofilm. This could be due to the different cluster expansion dynamics of the bacteria. For example, Staphylococcus spp. are non-motile and naturally arrange in clusters reflective of their inability to separate after division.36 Biofilm growth is then primarily based on clonal expansion that would exert outward pressure on the micropillars at the growing edges, but less at the focal points of growth. In contrast, E. coli and P. aeruginosa also utilize surface motility to facilitate the spreading of a developing biofilm and differentiation of a flat layer of cells.37–39
To further examine whether distinct deflection patterns generated by different species during biofilm growth could be detected, the alignment of deflection vectors at the edges of the biofilm were compared to the slope of the contour (blue line) using the cosine similarity (Fig. 5). In this case, deflection vectors that point in opposite directions (180° with respect to each other) but lie flat along the slope are considered parallel to the slope. Thus, cosine similarity values are absolute, with a θ value of 180° also giving a similarity of 1. In order of descending similarity and wider angles, the similarities and average angle between the deflection vector and contour were 0.69 ± 0.04 and 46.6 ± 3.5° for P. aeruginosa, 0.65 ± 0.01 and 49.8 ± 0.9° for E. coli, 0.59 ± 0.03 and 53.9 ± 2.1° for S. aureus, and 0.57 ± 0.05 and 55.1 ±3.7° for S. epidermidis, respectively. Thus, deflection vectors for the Staphylococcus spp. biofilms were less aligned to the biofilm contour, as compared to that of E. coli and P. aeruginosa biofilms, and this pattern may reflect different mechanisms of cluster expansion, i.e. clonal growth vs. surface motility.
Conclusions
There are limited tools available to study biofilm micromechanics and dynamics, with microscopy being one of the primary instruments of investigation. Microscopy is often combined with specialist techniques such as particle11 and cell-tracking,40 and usually requires high-end microscopes41 to probe biofilm dynamics. These techniques are labour intensive and time consuming. In addition, cell-tracking within mature biofilms is difficult because of the inaccuracy in resolving overlapping cells, and the long experimental times required due to slow cellular dynamics. Other micromechanical techniques used to investigate the physical properties of biological samples usually utilize equipment not available in biological laboratories. Examples of these include atomic force microscopy, optical light or laser traps, micropipette aspiration and magnetic tweezers. In this study, we present micropillar-embedded growth chambers as an experimental tool that can be used with standard, readily available, microscopes.Bacterial biofilms cause distinct deflection patterns of the micropillar arrays, which can be used to calculate differential pressure in biofilms, as shown here. The biomasses of the growing biofilms were physically constrained by, and exerted pressure on the micropillars. The heterogeneous development and differentiation of the biofilms resulted in differential pressure that was responsible for deflecting the micropillars. E. coli biofilms had the highest average differential pressure, and S. oneidensis biofilms the lowest. Although S. aureus and S. epidermidis biofilms had similar average differential pressures, the distribution and range of differential pressures experienced within their biofilms were different.
The generation of pressure differences that deflected the micropillars was dependent on EPS characteristics. In general, despite increases in biomass observed in some mutants, removal of any of the major EPS components strongly reduced the pressure differences within the biofilm, as measured here. EPS components that reduce stiffness and increase biofilm malleability are expected to enhance differential pressure, as they increase microbial dynamics, biofilm motion and lateral growth. In P. aeruginosa, the Psl polysaccharide has been shown to increase biofilm stiffness, whereas Pel is able to reduce biofilm stiffness and enhance overall malleability.11 In B. subtilis biofilms, mechanical forces are focused on areas of weakest matrix stiffness, causing differential pressure and buckling of the biofilm layer.8 EPS components that result in increased variation of biofilm mechanical properties are also expected to increase differential pressure. P. aeruginosa biofilms expressing Pel vary more in rheology spatially and temporally, as compared to Psl.11
The formation of high cell density areas and microcolonies in the strains tested was the major factor in disrupting the array and the creation of deflection patterns. Thus, micropillars may be sensitive to the different mechanisms that drive microcolony and cluster expansion for different bacteria. Micropillars may then be coupled with the use of specific mutants to probe, among other things, how EPS components or surface motility that affect biofilm dynamics and microcolony formation translate into pressure differences to affect biofilm morphology. It has been shown that pressure from biofilm expansion in confined spaces is responsible for the wrinkled structure of pellicles at the air–liquid interface.40 Differences in physical pressure generated by different bacterial strains may also have implications for the spatial organization of bacteria in multi-species biofilm communities. Such studies can help in our understanding of the mechanics of biofilm populations or communities.42
Finally, surfaces with high aspect ratio structures (tall in height and short in width), such as micropillars, have been employed in biofilm research and industry for their anti-biofouling properties.43 Thus, the methodology described here to measure differential pressure in biofilms can be coupled with such surfaces to produce a ‘smart’ material that detects when biofilms have eventually formed, thus having real application in the cleaning and maintenance of equipment in various industries.
Acknowledgements
We acknowledge financial support from the Singapore Centre for Environmental Life Sciences Engineering (SCELSE), whose research is funded by the National Research Foundation Singapore, Ministry of Education Singapore, Nanyang Technological University and National University of Singapore, under its Research Centre of Excellence program. We also acknowledge Singapore MIT Alliance for Research and Technology's research program in BioSystems and Micromechanics supported by the National Research Foundation Singapore.Notes and references
- H. C. Flemming and J. Wingender, Nat. Rev. Microbiol., 2010, 8, 623–633 CAS.
- J. W. Costerton, Z. Lewandowski, D. E. Caldwell, D. R. Korber and H. M. Lappin-Scott, Annu. Rev. Microbiol., 1995, 49, 711–745 CrossRef CAS PubMed.
- R. Singh, D. Paul and R. K. Jain, Trends Microbiol., 2006, 14, 389–397 CrossRef CAS PubMed.
- B. Q. Liao, D. M. Bagley, H. E. Kraemer, G. G. Leppard and S. N. Liss, Water Environ. Res., 2004, 76, 425–436 CrossRef CAS PubMed.
- P. S. Stewart and M. J. Franklin, Nat. Rev. Microbiol., 2008, 6, 199–210 CrossRef CAS PubMed.
- C. Douarche, J. M. Allain and E. Raspaud, Biophys. J., 2015, 109, 2195–2202 CrossRef CAS PubMed.
- A. Seminara, T. E. Angelini, J. N. Wilking, H. Vlamakis, S. Ebrahim, R. Kolter, D. A. Weitz and M. P. Brenner, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 1116–11121 CrossRef CAS PubMed.
- M. Asally, M. Kittisopikul, P. Rue, Y. J. Du, Z. X. Hu, T. Cagatay, A. B. Robinson, H. B. Lu, J. Garcia-Ojalvo and G. M. Suel, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 18891–18896 CrossRef CAS PubMed.
- A. K. Epstein, B. Pokroy, A. Seminara and J. Aizenberg, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 995–1000 CrossRef CAS PubMed.
- K. Drescher, Y. Shen, B. L. Bassler and H. A. Stone, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 4345–4350 CrossRef CAS PubMed.
- S. C. Chew, B. Kundukad, T. Seviour, J. R. C. van der Maarel, L. Yang, S. A. Rice, P. Doyle and S. Kjelleberg, mBio, 2014, 5, e01536-14 CrossRef PubMed.
- O. du Roure, A. Saez, A. Buguin, R. H. Austin, P. Chavrier, P. Silberzan and B. Ladoux, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 2390–2395 CrossRef CAS PubMed.
- M. T. Yang, J. P. Fu, Y. K. Wang, R. A. Desai and C. S. Chen, Nat. Protoc., 2011, 6, 187–213 CrossRef CAS PubMed.
- N. Biais, B. Ladoux, D. Higashi, M. So and M. Sheetz, PLoS Biol., 2008, 6, e87 Search PubMed.
- A. Cerf, J. C. Cau, C. Vieu and E. Dague, Langmuir, 2009, 25, 5731–5736 CrossRef CAS PubMed.
- H. H. P. Fang, K. Y. Chan and L. C. Xu, J. Microbiol. Methods, 2000, 40, 89–97 CrossRef CAS PubMed.
- S. Kesel, S. Grumbein, I. Gumperlein, M. Tallawi, A. K. Marel, O. Lieleg and M. Opitz, Appl. Environ. Microbiol., 2016, 82, 2424–2432 CrossRef PubMed.
- Y. Abe, P. Polyakov, S. Skali-Lami and G. Francius, Biofouling, 2011, 27, 739–750 CrossRef PubMed.
- S. M. Soto, A. Smithson, J. P. Horcajada, J. A. Martinez, J. P. Mensa and J. Vila, Clin. Microbiol. Infect., 2006, 12, 1034–1036 CrossRef CAS PubMed.
- J. R. Govan and V. Deretic, Microbiol. Rev., 1996, 60, 539–574 CAS.
- C. F. Schierle, M. De la Garza, T. A. Mustoe and R. D. Galiano, Wound Repair Regen., 2009, 17, 354–359 CrossRef PubMed.
- M. T. McCann, B. F. Gilmore and S. P. Gorman, J. Pharm. Pharmacol., 2008, 60, 1551–1571 CrossRef CAS PubMed.
- Y. A. Gorby, S. Yanina, J. S. McLean, K. M. Rosso, D. Moyles, A. Dohnalkova, T. J. Beveridge, I. S. Chang, B. H. Kim, K. S. Kim, D. E. Culley, S. B. Reed, M. F. Romine, D. A. Saffarini, E. A. Hill, L. Shi, D. A. Elias, D. W. Kennedy, G. Pinchuk, K. Watanabe, S. Ishii, B. Logan, K. H. Nealson and J. K. Fredrickson, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 11358–11363 CrossRef CAS PubMed.
- A. Folkesson, J. A. Haagensen, C. Zampaloni, C. Sternberg and S. Molin, PLoS One, 2008, 3, e1891 Search PubMed.
- H. Wu, B. Lee, L. Yang, H. Z. Wang, M. Givskov, S. Molin, N. Hoiby and Z. J. Song, FEMS Immunol. Med. Microbiol., 2011, 62, 49–56 CrossRef CAS PubMed.
- L. Yang, Y. Liu, T. Markussen, N. Hoiby, T. Tolker-Nielsen and S. Molin, FEMS Immunol. Med. Microbiol., 2011, 62, 339–347 CrossRef CAS PubMed.
- Z. Qin, Y. Ou, L. Yang, Y. Zhu, T. Tolker-Nielsen, S. Molin and D. Qu, Microbiology, 2007, 153, 2083–2092 CrossRef CAS PubMed.
- J. P. Fu, Y. K. Wang, M. T. Yang, R. A. Desai, X. A. Yu, Z. J. Liu and C. S. Chen, Nat. Methods, 2010, 7, U733–U795 CrossRef PubMed.
- A. Heydorn, A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll and S. Molin, Microbiology, 2000, 146, 2395–2407 CrossRef CAS PubMed.
- M. Vorregaard, Comstat2 - a modern 3D image analysis environment for biofilms, Technical University of Denmark, 2008 Search PubMed.
- J. M. Gere and S. P. Timoshenko, Mechanics of materials, PWS Publishing Co., Boston, 4th edn, 1997 Search PubMed.
- O. Galy, P. Latour-Lambert, K. Zrelli, J. M. Ghigo, C. Beloin and N. Henry, Biophys. J., 2012, 103, 1400–1408 CrossRef CAS PubMed.
- E. Oberg, Machinery's Handbook, Industrial Press, New York, 27th edn, 2005 Search PubMed.
- K. M. Colvin, Y. Irie, C. S. Tart, R. Urbano, J. C. Whitney, C. Ryder, P. L. Howell, D. J. Wozniak and M. R. Parsek, Environ. Microbiol., 2012, 14, 1913–1928 CrossRef CAS PubMed.
- K. C. Rice, E. E. Mann, J. L. Endres, E. C. Weiss, J. E. Cassat, M. S. Smeltzer and K. W. Bayles, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 8113–8118 CrossRef CAS PubMed.
- H. Tzagoloff and R. Novick, J. Bacteriol., 1977, 129, 343–350 CAS.
- M. Klausen, A. Aaes-Jorgensen, S. Molin and T. Tolker-Nielsen, Mol. Microbiol., 2003, 50, 61–68 CrossRef CAS PubMed.
- L. A. Pratt and R. Kolter, Mol. Microbiol., 1998, 30, 285–293 CrossRef CAS PubMed.
- G. A. O'Toole and R. Kolter, Mol. Microbiol., 1998, 30, 295–304 CrossRef.
- M. Trejo, C. Douarche, V. Bailleux, C. Poulard, S. Mariot, C. Regeard and E. Raspaud, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 2011–2016 CrossRef CAS PubMed.
- Z. Kun, T. Boo Shan, B. Bernard, J. Fan, L. G. Maxsim, J. H. Joe, L. Erik, R. P. Matthew and C. L. W. Gerard, Nature, 2013, 497, 388–391 CrossRef PubMed.
- A. Persat, C. D. Nadell, M. K. Kim, F. Ingremeau, A. Siryaporn, K. Drescher, N. S. Wingreen, B. L. Bassler, Z. Gitai and H. A. Stone, Cell, 2015, 161, 988–997 CrossRef CAS PubMed.
- A. K. Epstein, A. I. Hochbaum, P. Kim and J. Aizenberg, Nanotechnology, 2011, 22, 494007 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5sm02755a |
| This journal is © The Royal Society of Chemistry 2016 |




