Open Access Article
Open Access ArticleSquare channels formed by a peptide derived from transthyretin†
Stan
Yoo
,
Adam G.
Kreutzer
,
Nicholas L.
Truex
and
James S.
Nowick
*
Department of Chemistry, University of California, Irvine, California 92697-2025, USA. E-mail: jsnowick@uci.edu
First published on 1st August 2016
Abstract
High-resolution structures of peptide supramolecular assemblies are key to understanding amyloid diseases and designing peptide-based materials. This paper explores the supramolecular assembly of a macrocyclic β-sheet peptide derived from transthyretin (TTR). The peptide mimics the β-hairpin formed by the β-strands G and H of TTR, which form the interface of the TTR tetramer. X-ray crystallography reveals that the peptide does not form a tetramer, but rather assembles to form square channels. The square channels are formed by extended networks of β-sheets and pack in a “tilted windows” pattern. This unexpected structure represents an emergent property of the peptide and broadens the scope of known supramolecular assemblies of β-sheets.
Introduction
The structures of amyloid fibrils and oligomers represent a vast frontier, of yet unknown scope. The fibrils and aggregates that amyloidogenic peptides and proteins form are rich in β-sheets, and their structures are tremendously important in amyloid diseases. Many structures of amyloid fibrils have been discovered by solid-state NMR spectroscopy of amyloidogenic peptides and proteins and by X-ray crystallography of smaller fragments.1–4 Studying amyloid oligomer structures at high resolution is challenging, because amyloid oligomers are heterogeneous and dynamic, forming various species of different sizes and morphologies. Although a few structures of amyloid oligomers have been discovered in the last decade, there are not enough to provide a full understanding of amyloid assemblies.5–7Our laboratory has pioneered the use of macrocyclic β-sheets as a tool for exploring the structures of amyloid oligomers. In collaboration with the Eisenberg group, we began using X-ray crystallography to elucidate the oligomeric assemblies of macrocyclic β-sheets.8–10 Subsequently, our laboratory has independently elucidated oligomer structures formed by macrocyclic β-sheet peptides derived from Aβ, α-synuclein, and β2-microglobulin, which are associated with Alzheimer's disease, Parkinson's disease, and hemodialysis-related amyloidosis.11–13
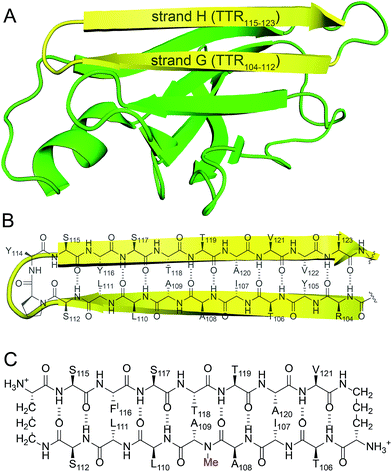
Transthyretin (TTR) is a tetrameric protein that is associated with amyloid diseases, including senile systemic amyloidosis and familial amyloid polyneuropathy.14 Two monomers of TTR come together in an edge-to-edge fashion to form dimers. The dimers further pack in a face-to-face fashion to form tetramers (Fig. 1).15 The TTR tetramers transport thyroid hormone and retinol-binding protein. Dissociation of the tetramers into monomers leads to formation of insoluble fibrils and toxic oligomers, which are associated with TTR-related amyloid diseases.16 Kelly and coworkers have invented an innovative approach for treating familial amyloid polyneuropathy by stabilizing the tetramers with a small molecule drug called tafamidis.17 Tafamidis acts by binding to the dimerization interface of two dimers and preventing dissociation into monomers.
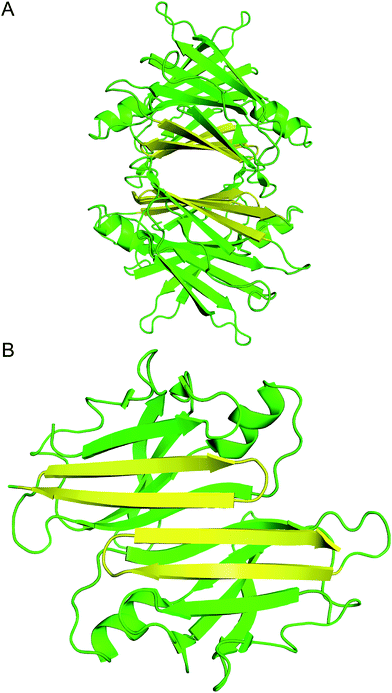 | ||
| Fig. 1 (A) X-ray crystallographic structure of the TTR tetramer. The β-hairpin of the tetramer interface is highlighted in yellow (PDB 1DVQ).18 (B) TTR dimer subunit. | ||
Inspired by the importance of the dimerization interface in stabilizing the TTR assembly and the propensity of TTR to form fibrils, we set out to elucidate the assembly of the β-hairpin that comprises the dimerization interface of the TTR tetramer.19 We incorporated the two β-strands from the TTR β-hairpin into a macrocyclic β-sheet and studied its assembly by X-ray crystallography. To our surprise, we discovered that the peptide exhibits a new mode of supramolecular assembly, forming square channels consisting of an extended network of β-sheets.
Results
Design, synthesis, and structure determination of peptide 1
We incorporated residues 106–112 from strand G of TTR and residues 115–121 from strand H into a macrocyclic β-sheet (Fig. 2). We used two δ-linked ornithine (δOrn) turn units to connect these two β-strands and form a macrocycle.20 We replaced the proline–tyrosine turn (residues 113 and 114) with a δOrn turn to promote β-hairpin formation. We connected residues 106 and 121 with another δOrn turn to reinforce the β-hairpin structure.21,22 We also incorporated an N-methyl group on alanine 109 (N-Me-A109) to prevent uncontrolled aggregation.23,24 We replaced tyrosine 116 with p-iodo-phenylalanine (FI116) to determine the X-ray crystallographic phases by single-wavelength anomalous diffraction (SAD) phasing.25Peptide 1 was synthesized as the trifluoroacetic acid (TFA) salt using standard Fmoc-based solid-phase peptide synthesis, solution-phase cyclization, and RP-HPLC purification. The TFA salt is readily soluble in water, but crystallizes or aggregates in buffer, depending on the conditions.26 Peptide 1 was screened in 288 crystallization conditions using Hampton Research crystallization kits (Crystal Screen, Index, and PEG/Ion). Peptide 1 grew rod-shaped crystals suitable for X-ray diffraction after two weeks in 0.1 M NaOAc buffer at pH 5.3, 0.2 M CaCl2, and 31% isopropanol. X-ray diffraction data were collected to 2.08 Å using an X-ray diffractometer with a Cu rotating anode. The X-ray crystallographic structure of peptide 1 was solved and refined in the P43212 space group, with one macrocycle in the asymmetric unit.
X-ray crystallography of peptide 1
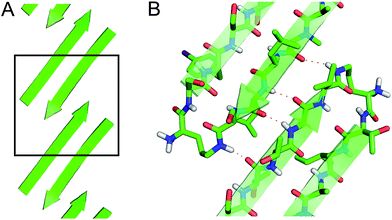
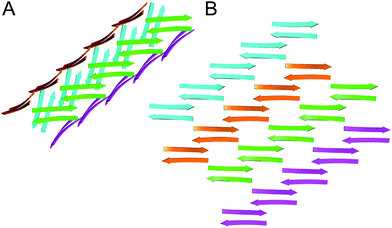
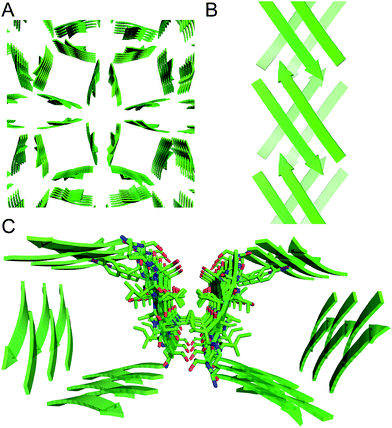
The X-ray crystallographic structure of peptide 1 reveals a hierarchical supramolecular assembly. The peptide folds to form a β-hairpin monomer. The β-hairpin assembles to form extended β-sheets. Four extended β-sheets form a square channel. The square channels pack into a pattern that resembles tilted windows.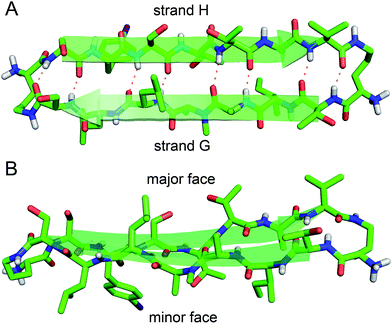 | ||
| Fig. 3 X-ray crystallographic structure of peptide 1 (PDB 5HPP). (A) Hydrogen-bonded β-hairpin monomer. (B) Side view of the β-hairpin monomer. | ||
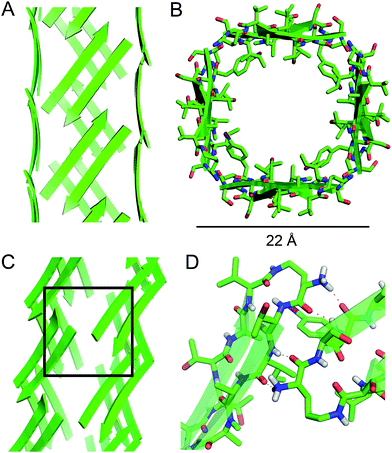 | ||
| Fig. 5 Square channel formed by peptide 1. (A) Side view. (B) Top view. (C) Corner view. (D) Detail showing hydrogen-bonding interactions between β-sheets at the corner of the square channel. | ||
Each square channel is stabilized by extended networks of hydrogen bonds between the four sides. The upper strand of each β-hairpin monomer in one side forms four hydrogen bonds with the lower strand of each β-hairpin monomer in the adjacent side (Fig. 5C and D). The amide oxygen of A108 hydrogen bonds with the α-NH of the ornithine that connects S112 and S115; the α-NH of I107 hydrogen bonds with the amide oxygen of S115; the amide oxygen of T106 hydrogen bonds with the α-NH of FI116; the α-NH of the ornithine that connects T106 and V121 hydrogen bonds with the amide oxygen of S117.
These two networks of hydrogen bonds—those within the extended β-sheets (Fig. 4) and those between the extended β-sheets (Fig. 5C and D)—stabilize the square channels. All of the amide NH and carbonyl groups participate in hydrogen bonding, except the α-NH of T118. To visualize how these two hydrogen-bonding networks act together to stabilize the square channels, one can imagine breaking all of the hydrogen bonds between two extended β-sheets at the corner of the square channel and unrolling and flattening the square channel (Fig. 6).
Discussion
In designing peptide 1 to mimic the TTR tetramerization interface, we anticipated that a tetramer would form (Fig. 8A). Instead of recapitulating the natural mode of assembly of TTR, peptide 1 forms a new mode of supramolecular assembly that has not been observed previously by X-ray crystallography—square channels. In the dimer subunit of the native TTR tetramer, strand H of one monomer pairs with strand H of another monomer. In the extended β-sheet formed by peptide 1, strand H pairs with strand G (Fig. 8B). The β-hairpins are shifted by two residues toward the N-terminus in the native TTR, while the β-hairpins in peptide 1 are shifted by four residues toward the C-terminus. The differences of the two assemblies may arise because other interactions, in addition to the β-sheet interactions, also stabilize the native TTR tetramer.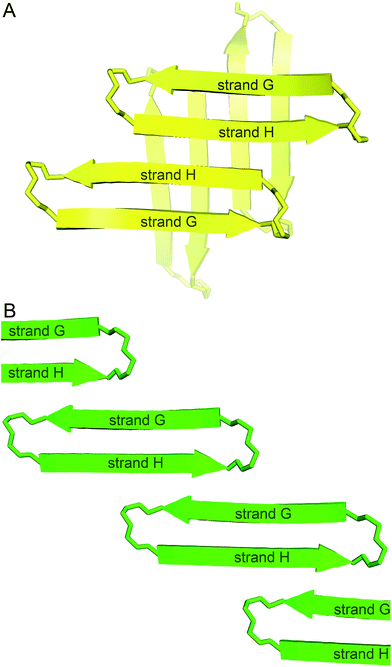 | ||
| Fig. 8 (A) Hypothesized tetramer of peptide 1. (B) Crystallographically observed extended β-sheet formed by peptide 1. | ||
The square channels formed by peptide 1 resemble β-barrels, because both are tubular structures that are composed of networks of hydrogen-bonded β-strands. Eisenberg and coworkers found that an amyloidogenic peptide from αB crystallin forms a six-stranded β-barrel, which they termed a “cylindrin” (Fig. 9).6 The square channels formed by peptide 1 resemble the cylindrin in that the interior is hydrophobic. The structures differ in that the square channels are hollow and run the length of the crystal, while the cylindrin forms a discrete hexamer with a tightly packed interior.
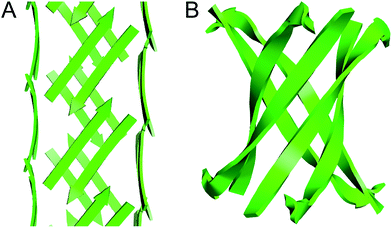 | ||
| Fig. 9 (A) Square channel formed by peptide 1. (B) Cylindrin formed by a peptide fragment from αB crystallin (PDB 3SGO). | ||
The interfaces between the square channels formed by peptide 1 resemble the layered β-sheet structures that compose amyloid fibrils. In amyloid fibrils, adjacent β-sheets form laminated layers in which the β-strands are nearly orthogonal to the fibril axis. The interfaces formed by the square channels in the “tilted windows” pattern are not as tightly laminated as many amyloid interfaces, such as the “steric zippers” discovered by Eisenberg and coworkers (Fig. 10).2 The β-strands of the square channels are not orthogonal to the channel axis, but rather are at about a 40° angle.
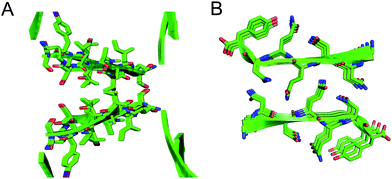 | ||
| Fig. 10 (A) Interface between square channels in the tilted windows assembly. (B) Steric zipper formed by a peptide from the yeast prion protein Sup35 (PDB 1YJP). | ||
Developing new peptide supramolecular assemblies enhances the understanding of peptide and protein interactions and also facilitates the creation of new molecular devices and functional materials.27–31 The supramolecular assemblies formed by peptide 1 are particularly reminiscent of a few important peptide nanostructures formed by β-sheets. Ghadiri and coworkers designed cyclic peptides that form hollow nanotubes in the solid state and in lipid bilayer membranes.32,33 The peptides are composed of alternating D- and L-amino acids and assemble to form extended β-sheet-like structures. Lynn and coworkers found that peptide fragments from the central region of Aβ, Aβ16–22, assemble to form large nanotubes composed of β-sheets.34,35 Schneider and coworkers developed peptide hydrogels as drug delivery vehicles.36–38 The hydrogels are composed of β-hairpin peptides with alternating hydrophobic and polar residues.
Although we do not yet understand all of the rules governing the supramolecular assembly of β-sheets, it is clear that the hydrogen-bonding edges impart Lego-like assembly.39 What makes the square channels formed by macrocyclic β-sheet peptide 1 especially interesting is that we did not design the peptide to assemble in this fashion, but rather this mode of assembly emerged from the structure and sequence of the peptide. The incorporation of a single N-methyl group helps reduce the near-infinite ways in which a pre-organized β-sheet can assemble and allows a single mode of self-assembly to emerge. As we continue to study the supramolecular assembly of β-sheet peptides containing different amino acid sequences, we anticipate better understanding how the structure and sequence of a peptide dictates its mode of supramolecular assembly.
Conclusion
In our efforts to explore the supramolecular assembly of β-sheets, we have designed a macrocyclic peptide derived from TTR. Rather than recapitulating the assembly of the native TTR tetramer, the peptide formed square channels that comprise extended networks of hydrogen-bonded β-sheets. This unanticipated result is intriguing, because the assembly represents an emergent property of the peptide. We anticipate that this new assembly of β-sheets will serve as a piece of puzzle toward understanding the rules of β-sheet supramolecular assembly.Acknowledgements
This work was supported by the National Science Foundation (NSF CHE-1507840).Notes and references
- A. T. Petkova, Y. Ishii, J. J. Balbach, O. N. Antzutkin, R. D. Leapman, F. Delaglio and R. Tycko, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 16742–16747 CrossRef CAS PubMed.
- R. Nelson, M. R. Sawaya, M. Balbirnie, A. O. Madsen, C. Riekel, R. Grothe and D. Eisenberg, Nature, 2005, 435, 773–778 CrossRef CAS PubMed.
- T. Luhrs, C. Ritter, M. Adrian, D. Riek-Loher, B. Bohrmann, H. Dobeli, D. Schubert and R. Riek, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 17342–17347 CrossRef CAS PubMed.
- M. R. Sawaya, S. Sambashivan, R. Nelson, M. I. Ivanova, S. A. Sievers, M. I. Apostol, M. J. Thompson, M. Balbirnie, J. J. Wiltzius, H. T. McFarlane, A. O. Madsen, C. Riekel and D. Eisenberg, Nature, 2007, 447, 453–457 CrossRef CAS PubMed.
- L. Yu, R. Edalji, J. E. Harlan, T. F. Holzman, A. P. Lopez, B. Labkovsky, H. Hillen, S. Barghorn, U. Ebert, P. L. Richardson, L. Miesbauer, L. Solomon, D. Bartley, K. Walter, R. W. Johnson, P. J. Hajduk and E. T. Olejniczak, Biochemistry, 2009, 48, 1870–1877 CrossRef CAS PubMed.
- A. Laganowsky, C. Liu, M. R. Sawaya, J. P. Whitelegge, J. Park, M. Zhao, A. Pensalfini, A. B. Soriaga, M. Landau, P. K. Teng, D. Cascio, C. Glabe and D. Eisenberg, Science, 2012, 335, 1228–1231 CrossRef CAS PubMed.
- M. I. Apostol, K. Perry and W. K. Surewicz, J. Am. Chem. Soc., 2013, 135, 10202–10205 CrossRef CAS PubMed.
- O. Khakshoor, A. J. Lin, T. P. Korman, M. R. Sawaya, S. C. Tsai, D. Eisenberg and J. S. Nowick, J. Am. Chem. Soc., 2010, 132, 11622–11628 CrossRef CAS PubMed.
- C. Liu, M. R. Sawaya, P. N. Cheng, J. Zheng, J. S. Nowick and D. Eisenberg, J. Am. Chem. Soc., 2011, 133, 6736–6744 CrossRef CAS PubMed.
- P. N. Cheng, C. Liu, M. Zhao, D. Eisenberg and J. S. Nowick, Nat. Chem., 2012, 4, 927–933 CrossRef CAS PubMed.
- A. G. Kreutzer, I. L. Hamza, R. K. Spencer and J. S. Nowick, J. Am. Chem. Soc., 2016, 138, 4634–4642 CrossRef CAS PubMed.
- P. J. Salveson, R. K. Spencer and J. S. Nowick, J. Am. Chem. Soc., 2016, 138, 4458–4467 CrossRef CAS PubMed.
- R. K. Spencer, A. G. Kreutzer, P. J. Salveson, H. Li and J. S. Nowick, J. Am. Chem. Soc., 2015, 137, 6304–6311 CrossRef CAS PubMed.
- C. Rapezzi, C. C. Quarta, L. Riva, S. Longhi, I. Gallelli, M. Lorenzini, P. Ciliberti, E. Biagini, F. Salvi and A. Branzi, Nat. Rev. Cardiol., 2010, 7, 398–408 CrossRef CAS PubMed.
- T. R. Foss, R. L. Wiseman and J. W. Kelly, Biochemistry, 2005, 44, 15525–15533 CrossRef CAS PubMed.
- K. Andersson, A. Olofsson, E. H. Nielsen, S.-E. Svehag and E. Lundgren, Biochem. Biophys. Res. Commun., 2002, 294, 309–314 CrossRef CAS PubMed.
- C. E. Bulawa, S. Connelly, M. Devit, L. Wang, C. Weigel, J. A. Fleming, J. Packman, E. T. Powers, R. L. Wiseman, T. R. Foss, I. A. Wilson, J. W. Kelly and R. Labaudiniere, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 9629–9634 CrossRef CAS PubMed.
- T. Klabunde, H. M. Petrassi, V. B. Oza, P. Raman, J. W. Kelly and J. C. Sacchettini, Nat. Struct. Biol., 2000, 7, 312–321 CrossRef CAS PubMed.
- A related cyclic peptide derived from the dimerization interface of the TTR tetramer has been found to inhibit Aβ aggregation: P. Y. Cho, G. Joshi, M. D. Boersma, J. A. Johnson and R. M. Murphy, ACS Chem. Neurosci., 2015, 6, 778–789 CrossRef CAS PubMed.
- J. S. Nowick and J. O. Brower, J. Am. Chem. Soc., 2003, 125, 876–877 CrossRef CAS PubMed.
- β-Strand G of TTR (105–115) forms amyloid fibrils and binds Aβ: (a) J. A. Jarvis, D. A. Craik and M. C. J. Wilce, Biochem. Biophys. Res. Commun., 1993, 192, 991–998 CrossRef CAS PubMed; (b) C. P. Jaroniec, C. E. MacPhee, N. S. Astrof, C. M. Dobson and R. G. Griffin, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 16748–16753 CrossRef CAS PubMed; (c) D. T. Yang, G. Joshi, P. Y. Cho, J. A. Johnson and R. M. Murphy, Biochemistry, 2013, 52, 2849–2861 CrossRef CAS PubMed.
- If this δOrn turn is omitted, the resulting acyclic peptide (H-Thr-Ile-Ala-N-Me-Ala-Leu-Leu-Ser-δOrn-Ser-PheI-Ser-Thr-Thr-Ala-Val-OH) does not crystallize under the conditions that peptide 1 crystallizes. It also does not aggregate in thioflavin T fluorescence assays.
- R. Spencer, K. H. Chen, G. Manuel and J. S. Nowick, Eur. J. Org. Chem., 2013, 3523–3528 CrossRef CAS.
- If the N-methyl group of peptide 1 is omitted, a gel forms when the cyclic peptide is deprotected. The placement of the N-methyl group on alanine 109 is important for crystallization; homologues with alternative placements of the N-methyl group do not crystallize under the conditions that peptide 1 crystallizes.
- R. K. Spencer and J. S. Nowick, Isr. J. Chem., 2015, 55, 698–710 CrossRef CAS PubMed.
- Thioflavin T fluorescence assays show that peptide 1 aggregates over the course of hours. Circular dichroism shows that the aggregates exhibit β-sheet structure. For details of these experiments, see the ESI.†.
- J. D. Hartgerink, E. Beniash and S. I. Stupp, Science, 2001, 294, 1684–1688 CrossRef CAS PubMed.
- V. Jayawarna, M. Ali, T. A. Jowitt, A. F. Miller, A. Saiani, J. E. Gough and R. V. Ulijn, Adv. Mater., 2006, 18, 611–614 CrossRef CAS.
- R. J. Swanekamp, J. T. DiMaio, C. J. Bowerman and B. L. Nilsson, J. Am. Chem. Soc., 2012, 134, 5556–5559 CrossRef CAS PubMed.
- M. Reches and E. Gazit, Nat. Nanotechnol., 2006, 1, 195–200 CrossRef CAS PubMed.
- B. Sanii, R. Kudirka, A. Cho, N. Venkateswaran, G. K. Olivier, A. M. Olson, H. Tran, R. M. Harada, L. Tan and R. N. Zuckermann, J. Am. Chem. Soc., 2011, 133, 20808–20815 CrossRef CAS PubMed.
- M. R. Ghadiri, J. R. Granja, R. A. Milligan, D. E. McRee and N. Khazanovich, Nature, 1993, 366, 324–327 CrossRef CAS PubMed.
- J. D. Hartgerink, J. R. Granja, R. A. Milligan and M. R. Ghadiri, J. Am. Chem. Soc., 1996, 118, 43–50 CrossRef CAS.
- K. Lu, J. Jacob, P. Thiyagarajan, V. P. Conticello and D. G. Lynn, J. Am. Chem. Soc., 2003, 125, 6391–6393 CrossRef CAS PubMed.
- A. K. Mehta, K. Lu, W. S. Childers, Y. Liang, S. N. Dublin, J. Dong, J. P. Snyder, S. V. Pingali, P. Thiyagarajan and D. G. Lynn, J. Am. Chem. Soc., 2008, 130, 9829–9835 CrossRef CAS PubMed.
- M. C. Branco, D. J. Pochan, N. J. Wagner and J. P. Schneider, Biomaterials, 2009, 30, 1339–1347 CrossRef CAS PubMed.
- J. E. Sun, B. Stewart, A. Litan, S. J. Lee, J. P. Schneider, S. A. Langhans and D. J. Pochan, Biomater. Sci., 2016, 4, 839–848 RSC.
- S. Lindsey, J. H. Piatt, P. Worthington, C. Sonmez, S. Satheye, J. P. Schneider, D. J. Pochan and S. A. Langhans, Biomacromolecules, 2015, 16, 2672–2683 CrossRef CAS PubMed.
- P. N. Cheng, J. D. Pham and J. S. Nowick, J. Am. Chem. Soc., 2013, 135, 5477–5492 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6sc01927g |
| This journal is © The Royal Society of Chemistry 2016 |