Open Access Article
Open Access ArticleTotal chemical synthesis of proteins without HPLC purification†
S. F.
Loibl
,
Z.
Harpaz‡
,
R.
Zitterbart‡
and
O.
Seitz
*
Humboldt-Universität zu Berlin, Institut für Chemie, Brook-Taylor-Str. 2, 12489 Berlin, Germany. E-mail: oliver.seitz@chemie.hu-berlin.de; Fax: +49 030 2093 7266; Tel: +49 030 2093 7446
First published on 4th July 2016
Abstract
The total chemical synthesis of proteins is a tedious and time-consuming endeavour. The typical steps involve solid phase synthesis of peptide thioesters and cysteinyl peptides, native chemical ligation (NCL) in solution, desulfurization or removal of ligation auxiliaries in the case of extended NCL as well as many intermediary and final HPLC purification steps. With an aim to facilitate and improve the throughput of protein synthesis we developed the first method for the rapid chemical total on-resin synthesis of proteins that proceeds without a single HPLC-purification step. The method relies on the combination of three orthogonal protein tags that allow sequential immobilization (via the N-terminal and C-terminal ends), extended native chemical ligation and release reactions. The peptide fragments to be ligated are prepared by conventional solid phase synthesis and used as crude materials in the subsequent steps. An N-terminal His6 unit permits selective immobilization of the full length peptide thioester onto Ni-NTA agarose beads. The C-terminal peptide fragment carries a C-terminal peptide hydrazide and an N-terminal 2-mercapto-2-phenyl-ethyl ligation auxiliary, which serves as a reactivity tag for the full length peptide. As a result, only full length peptides, not truncation products, react in the subsequent on-bead extended NCL. After auxiliary removal the ligation product is liberated into solution upon treatment with mild acid, and is concomitantly captured by an aldehyde-modified resin. This step allows the removal of the most frequently observed by-product in NCL chemistry, i.e. the hydrolysed peptide thioester (which does not contain a C-terminal peptide hydrazide). Finally, the target protein is released with diluted hydrazine or acid. We applied the method in the synthesis of 46 to 126 amino acid long MUC1 proteins comprising 2–6 copies of a 20mer tandem repeat sequence. Only three days were required for the parallel synthesis of 9 MUC1 proteins which were obtained in 8–33% overall yield with 90–98% purity despite the omission of HPLC purification.
Introduction
Solid phase peptide synthesis (SPPS)1 and native chemical ligation (NCL)2 are the key enabling methods used for the total chemical synthesis of proteins. Typically, up to 50 aa long peptide thioesters are prepared by SPPS and subsequently used in the native chemical ligation with cysteinyl peptides that have been, again, prepared by SPPS. Usually, the peptide fragments are purified by HPLC prior to NCL and HPLC purification is again required for isolation of the desired ligation product. However, HPLC purification is still a bottleneck to automation which limits throughput, upscaling and increases the costs of protein synthesis. On the other hand, recombinant synthesis of His6 or GST tagged proteins requires comparably little purification efforts once the overexpression system has been established, but cloning and cell culture is cumbersome.An ideal method would combine the ease of programmable solid-phase chemical synthesis with the effortless isolation procedures used in recombinant protein synthesis. One way to simplification of chemical protein synthesis is performing ligations on a solid support. Reagents or peptide fragments can be used in excess and can be readily removed by simple washing steps. The pioneering work from Kent and Dawson involved chemical linkages to sepharose and agarose type resins.3,4 The peptides were prepared by solid-phase peptide synthesis on conventional synthesis resins and purified by HPLC prior to solid-supported native chemical ligation. Brik performed the synthesis of a peptide as well as the subsequent native chemical ligation with HPLC purified peptides on a PEGA resin.5 Aucagne used the PEGA resin for immobilization of the N-terminal peptide via copper click chemistry, followed by iterative N-to-C directional elongation. The peptide fragments were, again, purified by HPLC prior to solid-supported NCL.6 The method has recently been extended to the use of non-purified peptides and was combined with iterative N-to-C-terminal copper-click and oxime ligation reactions, which provided up to 160 amino acid long glycopeptides containing three non-natural backbone substitutions.7–9
The use of resin capture-and-release methods has been considered as an alternative to solid-supported NCL. Kent connected the His6 tag to the C-terminal end of the C-terminal ligation fragment and used a Ni-NTA agarose resin to facilitate buffer exchanges after native chemical ligation in solution.10 Again, HPLC purification was involved in the peptide fragments to be ligated. Mende et al. and subsequently Zitterbart et al. were the first to introduce an HPLC-free total chemical synthesis of immobilized proteins.11,12 The methods relied on capture to microtiter plates by means of C-terminal tagging. However, the release of proteins into solution was not demonstrated and the purity of the protein remained unclear.
Despite the remarkable improvements, the HPLC-free total chemical synthesis of proteins in native and soluble form has not been described yet. The previous methods were restricted to cysteine-containing target proteins and therefore, the use of auxiliaries or ligation-desulfurization tactics – required to enable protein synthesis beyond cysteine containing ligation junctions – was not explored.
Results
Design
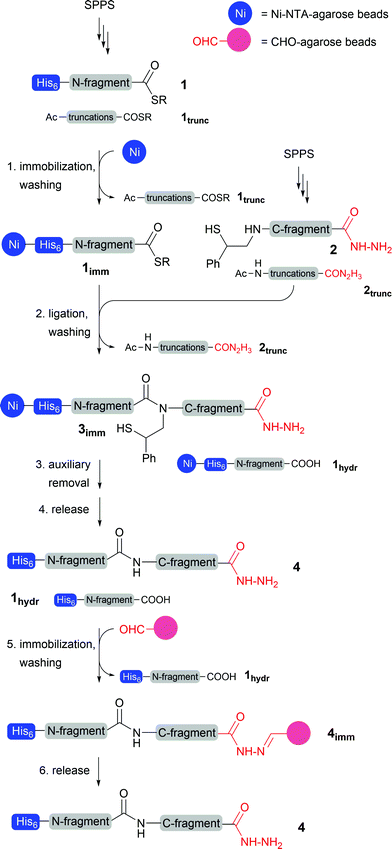
In the pursuit of a chemical protein synthesis method that bypasses the need for cysteine, is amenable to automation and avoids the arduous HPLC purification, we envisioned the combined use of a newly developed ligation auxiliary,13,14 two capture resins and solid-supported native chemical ligation (Scheme 1). In contrast to previous reports,10,11 we employed the His6 tag at the N-terminal rather than the C-terminal end of the nascent protein. His6 tagging of peptide thioesters is readily performed at the final stage of solid-phase synthesis. The His6 unit is routinely used in the synthesis of recombinant proteins.15 Importantly, the N-terminal His6 tag in 1 marks the selective immobilization of the His6-tagged thioesters 1 onto a Ni-NTA resin. After the removal of truncation products 1trunc by simple washing of the resin with aqueous buffer, the immobilized full-length peptide thioester 1imm is used in a solid-supported NCL reaction with the C-terminal fragment 2. The latter is obtained by Fmoc-based solid-phase synthesis. The synthesis of 2 features a reductive amination to introduce the 2-mercapto-2-phenylethyl auxiliary at the last step. This on the one hand extends the scope of solid-supported NCL chemistry beyond cysteine containing ligation junctions, and on the other hand provides the full-length C-terminal fragment 2 with a reactivity tag. Because capping will exclude the truncation products 2trunc from the introduction of the auxiliary, the NCL reaction can be performed using crudes. Subsequent washing of the Ni-NTA resin removes the truncation products 2trunc and yields the resin-bound ligation product 3imm. Cleavage of the 2-mercapto-2-phenethyl auxiliary with TCEP and morpholine provides the native peptide 4.The hydrolysis of peptide thioesters is the most frequently observed side reaction in NCL chemistry. Therefore, after solid-supported NCL the Ni-NTA resin carries both the ligation product 3imm and the hydrolysis product 1hydr(imm). For selective enrichment of the ligation product 4, a second capture reaction is designed to target the C-terminal end of the auxiliary peptide. The auxiliary-loaded peptides 2 are synthesized as peptide hydrazides, which were recently introduced for the Fmoc-based synthesis of peptide thioesters and as masked thioesters in sequential ligation reactions.16 We use the peptide hydrazide moiety as a purification tag which allows selective immobilization upon reaction with an aldehyde-functionalised and water-swellable agarose resin. The hydrazide-tag is easily introduced during SPPS and is significantly smaller than other established purification-tags, which should leave the protein’s functional properties unharmed. We saw a particular advantage in a sequence of capture reactions involving hydrazone ligation subsequent to Ni-His6 complexation. We planned to detach the peptides from the Ni-NTA resin by acidic treatment rather than the more commonly applied imidazole treatment at basic pH. Since hydrazones such as in 4imm are known to form rapidly under acidic conditions,17,18 the aldehyde-resin can be added directly to the cleavage solution without intermediate buffer exchange. In addition, acidic conditions help the solubilisation of many proteins. The capture step via hydrazone ligation allows the removal of the hydrolysis product 3hydr (by washing). As a last step treatment with diluted hydrazine liberated the ligation product 4, which will be obtained in high purity without HPLC purification.
The key advantage of this concept is that by using two orthogonal purification tags, a quantitative conversion of the immobilized thioester is not required anymore to yield the final product in high purity. Therefore, analysis of intermediates becomes redundant which should be especially useful for high-throughput chemical synthesis of proteins or protein-libraries.
To demonstrate the proof-of-feasibility for the HPLC-free protein synthesis we chose the MUC1 protein as a synthetic target. The extracellular domain of this protein consists of 20–125 tandem repeats (VNTR) of a 20 amino acid long sequence (Fig. 1A).19 The longest mucin accessed by total chemical synthesis contains 8 repeats, which are, however, interrupted by 3 artificial backbone linkages.6,7
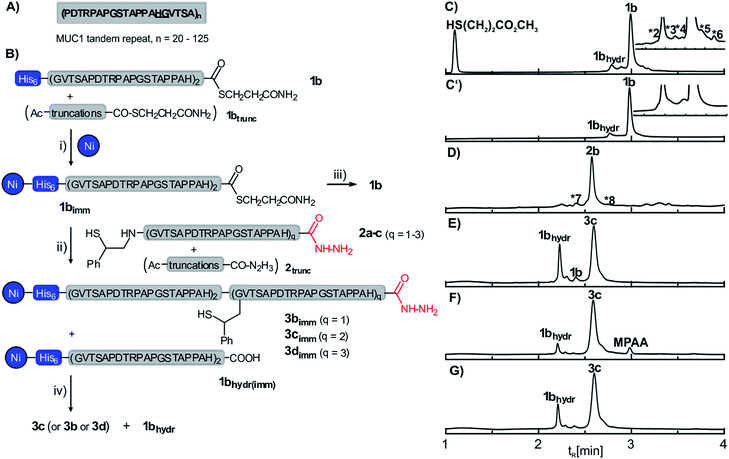 | ||
| Fig. 1 (A) Extracellular tandem repeat sequence of human mucin-1 (MUC1). The underlined residues mark the selected ligation junction. (B) Synthesis of MUC1 by solid-supported native chemical ligation: (i) 6 M GuHCl, 200 mM Na2HPO4, pH 7.5; (ii) 6 M GuHCl, 200 mM Na2HPO4, conditions given in Table 1; (iii) 0.1% aqueous TFA; (iv) 100 mM aqueous imidazole (pH 8.5). UPLC analysis (λ = 210 nm) of (C) crude peptide thioester 1b before and (C′) after capture-release from Ni-NTA agarose beads (*2: TAPPAH-COS(CH2)2COOCH3, *3: Ac-PAPGSTAPPAH-COS(CH2)2COOCH3, *4: full-length-COS(CH2)2COOH, *5: Ac-PAPGSTAPPAHGVTSAPDTRPAPGSTAPPAHCOS(CH2)2COOCH3, *6: unassigned); (D) crude auxiliary modified peptide hydrazide 2b (*7: H2N-(GVTSAPDTRPAPGSTAPPAH)2-CON2H3; *8: HS-Aux-HN-(GVTSAPDTRPAPGSTAPPAH)2-CON2HCOCF3); crude product obtained upon solid-supported NCL of 1b and 2b in the presence of (E) 20 mM TCEP, thiophenol, pH 7.0 (Table 1, entry 1); (F) 20 mM TCEP, MPAA, pH 7.5 (Table 1, entry 3); (G) 40 mM TCEP, 20 mM imidazole, thiophenol, pH 7.5 (Table 1, entry 4) (GuHCl, guanidinium hydrochloride; MPAA, mercaptophenylacetic acid; TCEP, triscarboxyethylphosphine; TFA, trifluoroacetic acid. HPLC conditions: detection at λ = 210 nm, gradients: 3–30% (B) in 4 min for (C)/(C′), 3–40% (B) in 4 min for (D–G); for details see ESI†). | ||
Synthesis
We embarked on the parallel synthesis of five MUC1 proteins consisting of 2 to 6 tandem repeats (or 46 to 126 amino acids including the His6 tag). The sequence of the VNTR-domain lacks a suitable cysteine residue for NCL but offers a His–Gly junction which is readily accessible by auxiliary-mediated peptide ligation. In the initial phase of the development we focused on the synthesis of a MUC1 protein comprising 4 tandem repeats (Fig. 1B). The His6-MUC1 peptide thioester 1b with a length of two tandem repeats was prepared on a chlorotrityl-resin by Fmoc-SPPS and subsequent in-solution thioesterification of the fully protected peptide acid in the presence of PyBOP, DIPEA and methyl 3-mercaptopropionate at −35 °C. UPLC analysis and co-elution studies with segments containing D-histidine at the C-terminus suggested that racemisation did not occur (Scheme S4†). The crude product obtained after TFA-treatment contained small amounts of truncation products (Fig. 1C). For immobilization, the crude peptide thioester was dissolved in buffer (6 M GuHCl, 200 mM Na2HPO4, pH 7.5) and added to the Ni-NTA agarose resin. A 15 min treatment was sufficient to quantitatively extract 1b. Subsequent washing of the resin with buffer and water led to the removal of the truncation products and afforded the resin-bound peptide thioester 1b in high purity (Fig. 1C′, see also Fig. S5†).For the C-terminal segment, we selected the 2-mercapto-2-phenethyl auxiliary (2b, Fig. 1B), which provides for efficient NCL reactions even at hindered sites.13 We anticipated fast reactions at the His–Gly junction due to the high reactivity of C-terminal His-peptide thioesters and the low impact of steric hindrance by auxiliaries at glycine. A preloaded Fmoc-His(Trt)-hydrazide chlorotrityl resin was elongated by means of automated Fmoc-SPPS. The 2-mercapto-2-phenethyl auxiliary was coupled onto the N-terminal Gly-residue by on-resin reductive amination. UPLC-MS analysis of the crude material obtained upon TFA-treatment revealed that high amounts of water (>8 vol%) were required to prevent trifluoracetylation of the peptide-hydrazide moiety (Fig. S9†).
Next we optimized the solid-supported NCL. The initial ligation experiments were performed using 2.5 equivalents auxiliary-loaded peptide 2b at pH 7 with TCEP and thiophenol as the NCL catalyst. After 24 h the resin-bound material was released by treatment with aqueous solution of imidazole (100 mM, pH 8.5) and analysed using UPLC-MS (Fig. 1E). The ligation product 3c was formed in 68% yield (Table 1). Of note, the truncation products from the solid-phase synthesis of fragments 1b and 2b (Fig. 1C and D) were absent, indicating the self-purification features of the approach. Slightly basic conditions (pH 7.5) helped to increase the yield to 77% (Table 1, entry 2). We considered the use of highly water soluble mercaptophenylacetic acid (MPAA). Indeed, replacement of thiophenol by MPAA allowed for further improvements of ligation yield (89%, Table 1 entry 3). However, it was difficult to remove MPAA during the subsequent washing steps (Fig. 1F). Considering the potential difficulties in the radical-triggered auxiliary cleavage, we therefore decided to keep thiophenol. Instead, we increased the amount of TCEP (from 20 mM to 40 mM) to exclude formation of disulfides and included imidazole (20 mM) to reduce potential unspecific interactions of the peptide fragments with the Ni-NTA resin, and the ligation product 3c was obtained in 81% yield (Fig. 1G). Please note, immobilization via the Ni·NTA·His6 complex remained stable under these conditions (Fig. S17b†). Next, we tested the robustness of the optimized SPCL conditions by performing the reaction between the immobilized 46mer peptide thioester 1bimm and the two 20mer and 60mer auxiliary-peptides 2a and 2c, respectively. Both SPCL reactions furnished the desired ligation products in ≥75% yield.
Since our approach is employing two orthogonal purification tags and thus quantitative yields in SPCL are not required in order to yield pure proteins, we next examined the removal of the auxiliary from the resin-bound ligation product (Fig. 2A). Treatment of the auxiliary-containing ligation product on the resin (3c) with an aqueous solution (pH 8.5) of TCEP (200 mM) and morpholine (800 mM) at 40 °C triggered the desired auxiliary cleavage but also caused partial detachment of the His6-tagged peptides, which is probably due to the high concentration of phosphine. A simple adjustment of the aqueous reaction mixture to pH 7.0 by addition of 1 M HCl led to reimmobilization of the His6-tagged peptides, which simplified the removal of auxiliary debris and excess reagent by washing (see also Fig. S17†). UPLC-MS analysis showed a minor amount of a peptidyl morpholine adduct which presumably was formed in addition to the hydrolysis product 1bhydr from the remaining non-hydrolyzed peptide thioester 1b (Fig. 2B). In the next step, the ligation product 4c (as well as the His6-tagged peptide acid 1bhydr) was released from the resin by treatment with 0.25 M acetic acid. The acidic filtrate was added to an aldehyde-functionalized agarose resin. Only the ligation product 4c contains a C-terminal hydrazide which readily forms the hydrazone 4cimm2 under the acidic conditions. The resin-captured ligation product was washed with GuHCl containing buffer to remove the His6-tagged peptide acid 1bhydr (Fig. 2C). Finally, the resin 4cimm2 was treated with 0.5 vol% of hydrazine in water, which furnished the 86 aa MUC1 protein 4c in 21% yield (based on initial loading of the SPPS-resin) at high apparent purity (>90%, see S11, ESI†).
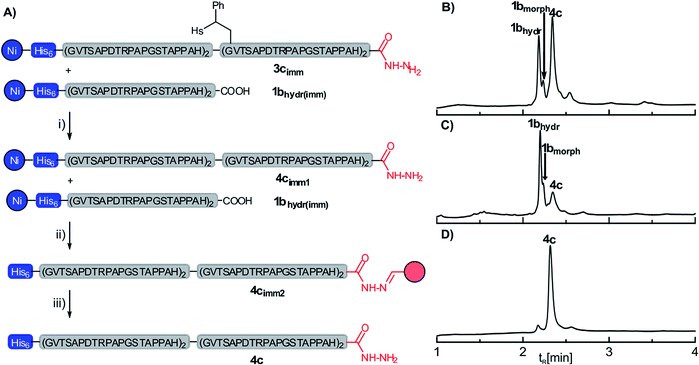 | ||
| Fig. 2 (A) Solid-supported removal of the ligation auxiliary from the ligation product 3cimm and separation from the hydrolysis product 1bhydrvia capture-and-release by means of aldehyde-functionalized agarose beads (red); (i) 0.2 M TCEP, 0.8 M morpholine, 24 h, r.t.; (ii) 0.25 M acetic acid; (iii) 0.5 vol% hydrazine. UPLC analysis of (B) the crude product obtained after the removal of the ligation auxiliary (step i); (C) eluates after washing of 4cimm2 and (D) the final crude product after steps (i), (ii) and (iii). HPLC conditions: detection at λ = 210 nm, for details see ESI.† | ||
We applied the method in the synthesis of 8 additional MUC1 peptides. Three different peptide thioesters (1a–c, for synthesis see the ESI†) containing 26, 46 or 66 amino acids and three different auxiliary-armed peptides 2a–c (20, 40 and 60 amino acids) were used in 8 additional solid-supported NCL reactions (Fig. 3A). A parallel workflow enabled the rapid synthesis of five MUC1 proteins consisting of 46 to 126 amino acids (46 to 126 amino acids). According to UPLC-MS analysis the 9 crude products obtained in the parallel 5-step synthesis, i.e. (i) immobilization to Ni–agarose, (ii) NCL, (iii) removal of the ligation auxiliary, (iv) release from Ni–agarose and concomitant capture with aldehyde–agarose, and (v) final release, were obtained in 90-98% apparent purity (λ = 210 nm) (Fig. 3B–G) and isolated yields between 8 and 33%. This level of purity should be sufficient for investigations of biological properties. HPLC purification may in principle provide even higher purities. However, we noticed that some separation problems are not in the reach of HPLC (see for example synthesis of 86 aa long 4c, Fig. S16†). In such cases, separation based on chemical reactivity (e.g. His tags and peptide hydrazide) will be advantageous.
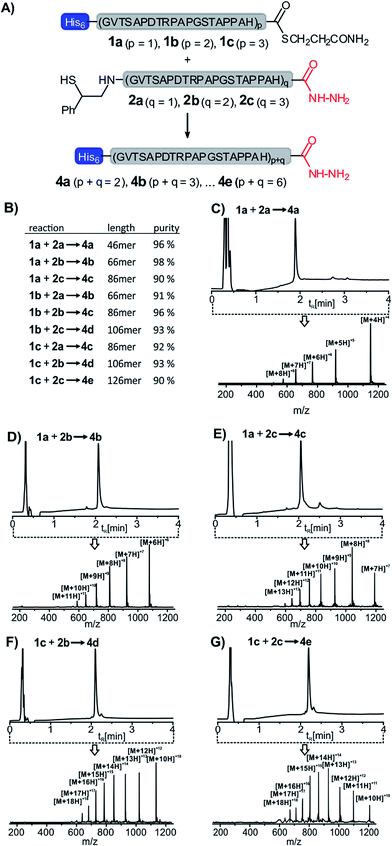 | ||
| Fig. 3 (A) Solid-supported synthesis of MUC1 proteins 4a–e as described in Scheme 1. (B) Purities of the crude ligation products and (C)–(G) UPLC-MS analyses of exemplary products. HPLC conditions: see Fig. 1. ESI-mass spectra were recorded in mass range (m/z): 150–1250 and integrated over the entire run time. | ||
Discussion
The repeated immobilization, washing and release steps may hint to a potential drawback of the method. The reactions used for immobilization and release may not succeed quantitatively and given the reversibility of immobilization, losses may also occur during washing. Indeed, we noticed that the hydrazone formed upon capture of peptide hydrazides with the aldehyde–agarose is not stable enough to completely avoid leakage during washing (see Fig. 2C). Yet, it should be considered that HPLC purification also leads to losses of peptide material (approx. 30% per run). The amount of MUC1 proteins 4a–e isolated after solid phase peptide synthesis and the 5 step protein assembly (Scheme 1) corresponds to 8–33% yield. Given that the calculation is based on the initial histidine load of the chlorotrityl resin used for synthesis of the peptide thioester, we conclude that the HPLC purification-free protein synthesis method can proceed as efficiently as conventional peptide ligation methods relying on solution-phase chemistry and HPLC purification. Furthermore, the avoidance of HPLC purification should facilitate upscaling.Most steps were performed under denaturing conditions (6 M guanidinium hydrochloride during ligation and washing, AcOH for cleavage of the Ni·His6 linkage and subsequent immobilization onto the aldehyde-functionalized agarose beads) which are required for solubilisation of many proteins and which may, in cases, be necessary to prevent aggregation on-resin. Though protein solubility is not an issue with MUC1 proteins, it would add to the usefulness of the method if denaturing conditions were also applicable for auxiliary cleavage and final release from the aldehyde resin. We have previously shown that auxiliary removal also proceeds in the presence of 6 M GuHCl.13 Furthermore we showed that hydrazine cleavage can also be accomplished by means of 1% trifluoroacetic acid (Fig. S18†). In contrast to GnHCl, this denaturing agent is readily removed upon lyophilisation.
One of the main drawbacks of bead-supported native chemical ligation is the reaction rate. Though we have not analysed reaction rates thoroughly, it is obvious that the solid-supported NCL investigated here proceeded slower (24 h reaction time) than expected for solution phase reactions. The NCL rates are probably hampered due to the slow diffusion of the peptide segments through the polymeric network. Given the high efficiency of NCL on surfaces,12 it is plausible to assume that increases in the rate of solid-supported NCL can be achieved when non-swellable, large surface beads are used.
Regardless of the reactivity issues discussed above, it is the reduction of time and expenses that is the most important hallmark in favour of our HPLC-purification-free protein synthesis. Of note, after solid-phase synthesis of the fragments, the preparation and capture-release based purification of the 9 MUC1 proteins was achieved in only three days. At the scale (0.5 μmol) used by us, less than 100 ml of aqueous waste was produced. In comparison, a HPLC-purification based synthesis of nine proteins would have consumed around 60-fold more HPLC-grade solvents and would have required orders of magnitude more time. The saving of time will become even more important in synthesis campaigns aiming for a larger number of proteins. Please note, the applied steps involve liquid handling and filtration. Intermediary lyophilisation is not required and therefore the process should be amenable to automation. While we focused on the use of the recently developed 2-mercapto-2-phenylethyl auxiliary, we wish to note that the method should also be applicable to the more commonly used ligation–desulfurisation tactics.20–22 With the dual tagging approach proposed by us, the number of different protein targets will only be limited by the throughput of the synthesis and dispensing robots. This may open an avenue to the parallel exploration of posttranslational protein modification.
Conclusions
In conclusion we have introduced the first method for the rapid chemical total synthesis of proteins without HPLC-purification. The method relies on extended native chemical ligation between peptide thioesters (N-terminal peptide fragment) and C-terminal peptide fragments bearing a recently developed 2-mercapto-2-phenyl-ethyl ligation auxiliary. Both peptide segments are prepared by conventional solid phase synthesis and are used as crude materials in the subsequent steps. The combination of an N-terminal His6 tag at the peptide thioester with the N-terminal auxiliary and the C-terminal hydrazide tag at the C-terminal peptide fragment is enabled for three major reasons: (1) native chemical ligation can be performed on agarose beads and washing facilitates the removal of buffer salts and additives used to drive NCL chemistry, (2) the N-terminal tags (His6 and ligation auxiliary) exclude solid-phase synthesis truncation products from NCL chemistry and (3) the C-terminal acyl hydrazide tag allows selective extraction of the ligation product. These thereby facilitate the removal of the most frequently observed NCL by-product, i.e. the hydrolysed peptide thioester. We demonstrated the method in the synthesis of 46 to 126 aa long MUC1 proteins. Only three days were required for the parallel synthesis of 9 MUC1 proteins which were obtained in 8–33% yield with 90–98% purity despite the omission of HPLC purification. We anticipate that our method will be helpful for building up large protein libraries in the future.Acknowledgements
We acknowledge support from Deutsche Forschungsgemeinschaft (Se819/15-1, SPP 1623).Notes and references
- R. B. Merrifield, J. Am. Chem. Soc., 1963, 85, 2149–2154 CrossRef CAS.
- P. Dawson, T. Muir, I. Clark-Lewis and S. Kent, Science, 1994, 266, 776–779 CAS.
- A. Brik, E. Keinan and P. E. Dawson, J. Org. Chem., 2000, 65, 3829–3835 CrossRef CAS PubMed.
- L. E. Canne, P. Botti, R. J. Simon, Y. Chen, E. A. Dennis and S. B. H. Kent, J. Am. Chem. Soc., 1999, 121, 8720–8727 CrossRef CAS.
- M. Jbara, M. Seenaiah and A. Brik, Chem. Commun., 2014, 50, 12534–12537 RSC.
- V. Aucagne, I. E. Valverde, P. Marceau, M. Galibert, N. Dendane and A. F. Delmas, Angew. Chem., Int. Ed., 2012, 51, 11320–11324 CrossRef CAS PubMed.
- M. Galibert, V. Piller, F. Piller, V. Aucagne and A. F. Delmas, Chem. Sci., 2015, 6, 3617–3623 RSC.
- I. E. Decostaire, D. Lelievre, V. Aucagne and A. F. Delmas, Org. Biomol. Chem., 2014, 12, 5536–5543 CAS.
- L. Raibaut, H. Adihou, R. Desmet, A. F. Delmas, V. Aucagne and O. Melnyk, Chem. Sci., 2013, 4, 4061–4066 RSC.
- D. Bang and S. B. H. Kent, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 5014–5019 CrossRef CAS PubMed.
- F. Mende, M. Beisswenger and O. Seitz, J. Am. Chem. Soc., 2010, 132, 11110–11118 CrossRef CAS PubMed.
- R. Zitterbart and O. Seitz, Angew. Chem., Int. Ed., 2016, 55, 7252–7256 CrossRef CAS PubMed.
- S. F. Loibl, Z. Harpaz and O. Seitz, Angew. Chem., Int. Ed., 2015, 54, 15055–15059 CrossRef CAS PubMed.
- Z. Harpaz, S. Loibl and O. Seitz, Bioorg. Med. Chem. Lett., 2016, 26, 1434–1437 CrossRef CAS PubMed.
- E. Hochuli, W. Bannwarth, H. Dobeli, R. Gentz and D. Stuber, Nat. Biotechnol., 1988, 6, 1321–1325 CrossRef CAS.
- G.-M. Fang, Y.-M. Li, F. Shen, Y.-C. Huang, J.-B. Li, Y. Lin, H.-K. Cui and L. Liu, Angew. Chem., Int. Ed., 2011, 50, 7645–7649 CrossRef CAS PubMed.
- A. Dirksen and P. E. Dawson, Bioconjugate Chem., 2008, 19, 2543–2548 CrossRef CAS PubMed.
- A. Dirksen, S. Dirksen, T. M. Hackeng and P. E. Dawson, J. Am. Chem. Soc., 2006, 128, 15602–15603 CrossRef CAS PubMed.
- J. Taylor-Papadimitriou, J. Burchell, D. W. Miles and M. Dalziel, Biochim. Biophys. Acta, 1999, 1455, 301–313 CrossRef CAS.
- L. Z. Yan and P. E. Dawson, J. Am. Chem. Soc., 2001, 123, 526–533 CrossRef CAS PubMed.
- H. Rohde and O. Seitz, Pept. Sci., 2010, 94, 551–559 CrossRef CAS PubMed.
- L. R. Malins and R. J. Payne, Aust. J. Chem., 2015, 68, 521–537 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6sc01883a |
| ‡ Contributed equally. |
| This journal is © The Royal Society of Chemistry 2016 |