Open Access Article
Open Access ArticleImprovement of enzyme-linked immunosorbent assay for the multicolor detection of biomarkers†
Chao
Li
a,
Yucai
Yang
b,
Dan
Wu
a,
Tianqi
Li
a,
Yongmei
Yin
b and
Genxi
Li
*ac
aState Key Laboratory of Pharmaceutical Biotechnology, Collaborative Innovation Center of Chemistry for Life Sciences, Department of Biochemistry, Nanjing University, Nanjing 210093, China. E-mail: genxili@nju.edu.cn
bDepartment of Oncology, The First Affiliated Hospital of Nanjing Medical University, Nanjing 210029, China
cLaboratory of Biosensing Technology, School of Life Sciences, Shanghai University, Shanghai 200444, China
First published on 3rd February 2016
Abstract
An enzyme-linked immunosorbent assay that is dependent on enzyme amplification has dominated the current field of protein detection; however, limited multiple detection ability and susceptible enzymatic reactions, and low sensitivity may severely hinder its application. Here, we report a new signal amplification scheme based on allochroic molecule modified carboxyl graphene oxide (cGO), which can be used to develop a multicolor immunoassay named as allochroic-cGO linked immunosorbent assay (ALISA). Thanks to high adsorption levels and a wide selection of allochroic molecules, the simultaneous colorimetric detection of diagnostic biomarkers at a picogram level can be successfully achieved for the first time. In addition, the color change triggered by acidic or basic water can provide a simple, rapid, stable and economical signal output, further meeting the growing biodetection requirements. Moreover, with the help of ALISA, we demonstrate that the combined detection of three tumor biomarkers, including carcino-embryonic antigen, neuron-specific enolase, and cytokeratin-19 fragment, is more valuable for differentiating lung cancer patients than the detection of a single biomarker, further manifesting the superiority of ALISA. All in all, this straightforward approach not only opens up new prospects for multicolor immunoassays, but also has great potential for applications in resource-constrained settings.
Introduction
The rapid, simultaneous, and sensitive detection of proteins associated with human diseases plays a fundamental role in clinical diagnosis, treatment and outcome evaluation. At present, enzyme-linked immunosorbent assay (ELISA), without a doubt, dominates the field of protein quantitation. Generally, ELISA is a technique that is based on the highly specific molecular recognition of antibody with the epitope of an antigen and an enzyme, such as horseradish peroxidase (HRP), used as catalytic labels for visual detection.1,2 Despite its widespread use, some limitations have to be comprehensively considered in the practical application. Firstly, the multiplex detection ability of ELISA is seriously deficient because of the simplex enzyme/substrate reaction, whereas a single marker, such as carcino-embryonic antigen (CEA), is often inadequate for the diagnosis of heterogeneous and complex diseases like cancer.3,4 Secondly, most of the commercial ELISA kits need targets in nanogram concentrations for naked-eye signal readout, whereas the concentration of many disease-related biomarkers in biological media is so low that they cannot be clearly identified without the help of a plate reader.5,6 Thirdly, the nonlinear nature of enzymatic amplification often encounters many problems including long time incubation, endogenous peroxide interference, mutagenic or carcinogenic substrates, and activity reduction resulting from preservatives or metal contamination.7 Therefore, it is urgent to program and implement a new signal amplification scheme that circumvents the above mentioned limitations of conventional ELISA.As a matter of fact, many efforts have been devoted to overcoming the shortcomings of conventional ELISA. For example, the integration of immunoassay with different kinds of nanotags such as DNA barcodes,8 semiconductor quantum dots,9 metal nanoparticles,10,11 and artificial antibody12 has been a popular way to detect multiple protein targets simultaneously. These methods successfully resolve one of the major challenges in multiplex detection, i.e., to identify each specific reaction with a distinct signal output. However, in these improved ELISA systems, the original colorimetric readout has to be changed to fluorescence, electrochemical, or magnetoresistance, which cannot be directly adapted to the conventional ELISA system, thus the advantages of ELISA have also been impaired. To the best of our knowledge, the multiplex colorimetric detection of proteins based on the existing ELISA systems has not been successfully explored until now.
Another example for the improvement of ELISA is the development of gold nanoparticle (AuNP)-based plasmonic sensors to analyze targets at ultralow concentrations, so as to enhance the sensitivity of ELISA.13,14 Although the AuNP-based strategy can be directly imposed on conventional ELISA, unfortunately, the dependence of enzyme-based amplification makes it almost impossible to implement multiplex biosensing.
Herein, for the first time, we report a novel signal amplification scheme composed of small allochroic dyes and antibody modified carboxyl graphene oxide (cGO) for the simple, sensitive, and multicolor detection of disease-related proteins. This improved ELISA, named as ALISA, i.e., allochroic-cGO linked immunosorbent assay, is based on the rapid release of chromatic dyes that are tightly bound to the surface of cGO, upon the simple addition of acidic or basic water (AW or BW) to the test solution. The chromatic dyes used here are organic compounds, such as malachite green carbinol base (MGCB),15 that will cause the color of the solution to change depending on the pH of the test solution. In this study, the above mentioned problems in improving ELISA have been well addressed, because (1) the detection sensitivity is expected to be considerably enhanced owing to the fact that large amounts of adsorbed dye molecules on the cGO surface through π–π stacking interactions can be released into the solution and lead to a strong color change; (2) different allochroic molecules are available to act as colorful barcodes resulting in the possibility of multicolor detection; (3) the signal output is fully determined from the total number of chromatic dyes bound to cGO, which ensures simple, rapid, stable and tunable colorimetric assays.
Results and discussion
As a proof of concept, we first prepared a functionalized nanoprobe composed of detection antibodies (denoted as Ab2), MGCB molecules, and a cGO nanosheet using a step-by-step procedure (Scheme 1). In this study, we make use of cGO instead of GO to immobilize more recognition molecules and to avoid irreversible agglomeration during the immobilization of the dyes and antibodies. cGO can be easily conjugated with Ab2 via the formation of a carbodiimide using N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide (EDC) as a coupling agent and N-hydroxysuccinimide (NHS) as an activator.16 After adding bovine serum albumin (BSA) to block the residual NHS moiety, we dropwise titrated the synthesized Ab2–cGO with MGCB in dimethyl sulfoxide because of the insolubility of MGCB in neutral aqueous solution. The MGCB–Ab2–cGO conjugates are then separated from the reaction solution through a centrifugation process, and characterized using physical and chemical techniques. Later, this MGCB–Ab2–cGO format can specifically recognize the analyte that has been captured by the capture antibody (denoted as Ab1). Upon the introduction of AW into the detection system, the attached MGCB molecules transform into soluble MG cations and are rapidly removed from the cGO surface, leading to the significant recovery of color readout.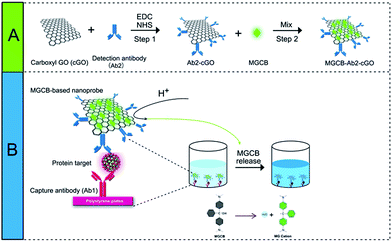 | ||
| Scheme 1 Schematic representation of the allochroic-cGO linked immunosorbent assay (ALISA) for protein detection performed in 96-well polystyrene (PS) plates. | ||
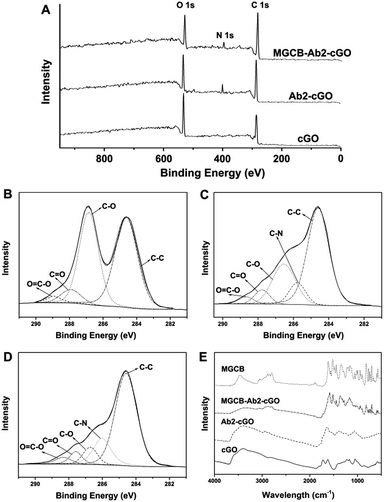
Fig. 1A shows the spectra from the X-ray photoelectron spectroscopic (XPS) measurements of cGO, Ab2–cGO and cGO–MGCB–Ab2. As expected, the spectrum of cGO shows only C and O peaks. Compared with cGO, the spectrum of Ab2–cGO shows the presence of N 1 s (398.8 eV) and a decrease in the intensity ratio of the O 1 s to the C 1 s peak, which indicates that the functionalization of cGO by antibody successfully occurred. The O content drops from 32.8% for cGO to 24.1% for Ab2–cGO owing to the reaction between the carboxyl groups of cGO and the amine group of Ab2. After further incubating with triphenyl-containing MGCB, the significant decrease of the O/C atomic abundance ratio from 0.363 for Ab2–cGO to 0.265 for MGCB–Ab2–cGO indicates a large amout of MGCB adsorption. Furthermore, the deconvolution of the C 1 s spectrum of cGO (Fig. 1B) points to the presence of four kinds of carbon atoms in different functional groups: C–C (ring carbons, 284.6 eV), C–O (carbons in phenolic hydroxyl groups, 286.7 eV), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (carbonyl, 287.8 eV), and O
O (carbonyl, 287.8 eV), and O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–OH (carboxylic, 288.8 eV). Of note, the intensities of the peaks related to the oxidized carbon species are greatly weakened upon amidation (Fig. 1C). Meanwhile, the new C–N species at approximately 286.1 eV in the C 1s spectrum of Ab2–cGO is attributed to the C–N bond between cGO and antibody as well as the C–N bonds in the protein molecules.17 After the introduction of MGCB, the peaks associated with C–C or C–H (284.6 eV) become more prominent, owing to the abundant phenyl rings of the MGCB molecules (Fig. 1D).
C–OH (carboxylic, 288.8 eV). Of note, the intensities of the peaks related to the oxidized carbon species are greatly weakened upon amidation (Fig. 1C). Meanwhile, the new C–N species at approximately 286.1 eV in the C 1s spectrum of Ab2–cGO is attributed to the C–N bond between cGO and antibody as well as the C–N bonds in the protein molecules.17 After the introduction of MGCB, the peaks associated with C–C or C–H (284.6 eV) become more prominent, owing to the abundant phenyl rings of the MGCB molecules (Fig. 1D).
The functionalized nanomaterial prepared has also been characterized using Fourier transform infrared spectroscopy (FTIR), and the spectra of cGO, Ab2–cGO, MGCB–Ab2–cGO, and MGCB are shown in Fig. 1E. The spectrum of cGO shows the characteristic peaks at 3420 (C–OH), 1730 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1628 (C
O), 1628 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), and 1058 cm−1 (C–O). After conjugation of Ab2, the peak at 1730 cm−1 almost disappears and obvious vibration peaks emerge corresponding to the featured amide bands of the protein [e.g., amide I (1645 cm−1) for C
C), and 1058 cm−1 (C–O). After conjugation of Ab2, the peak at 1730 cm−1 almost disappears and obvious vibration peaks emerge corresponding to the featured amide bands of the protein [e.g., amide I (1645 cm−1) for C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching and amide II (1539 cm−1) for N–H bending/C–N stretching], which confirms that the antibody has been successfully linked with cGO through the amidation reaction and the secondary structures of the immobilized antibody are fully retained.18 Compared with the FTIR spectra of MGCB and Ab2–cGO, the FTIR spectrum of MGCB–Ab2–cGO shows a similar fingerprint region to MGCB (1500–500 cm−1), which strongly suggests that MGCB is successfully attached to the Ab2–cGO nanosheets.
O stretching and amide II (1539 cm−1) for N–H bending/C–N stretching], which confirms that the antibody has been successfully linked with cGO through the amidation reaction and the secondary structures of the immobilized antibody are fully retained.18 Compared with the FTIR spectra of MGCB and Ab2–cGO, the FTIR spectrum of MGCB–Ab2–cGO shows a similar fingerprint region to MGCB (1500–500 cm−1), which strongly suggests that MGCB is successfully attached to the Ab2–cGO nanosheets.
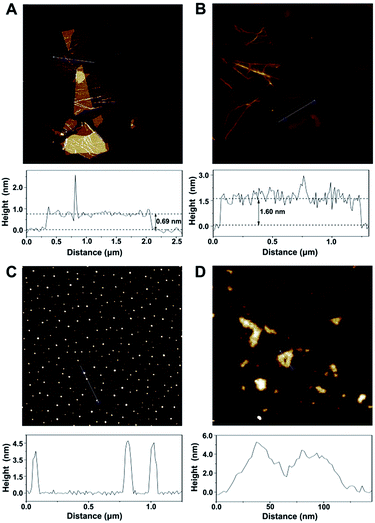
According to the topographical profiles analyzed using atomic force microscopy (Fig. 2), the height of the bare cGO sheets is approximately 0.69 nm, which confirms a monolayer state for the graphene sheets,19 while the average thickness of cGO-MGCB is determined to be about 1.60 nm. There is a 0.91 nm increment and an increased surface roughness compared with that of unmodified cGO, owing to the presence of MGCB on the graphene sheet surfaces. Since MGCB can attach on both sides of cGO, the thickness of the MGCB layer on graphene is calculated to be about 0.455 nm. This data is consistent with the thickness of a benzene derivative (0.47 nm).20 After conjugation of Ab2 (∼3.5 nm), the MGCB–Ab2–cGO nanosheets exhibit well-distributed brighter spots with increased heights (∼5.0 nm) on their surfaces, which may explain the dramatically enhanced dispersibility of the complex in aqueous media on account of the steric hindrance. Moreover, because the polydisperse (submicrometer to tens of micrometers) nature of the original cGO is harmful to the performance of the immunoassay, ultrasonic treatment has been used to break large nanosheets into nanometer cGO (Fig. 2D).
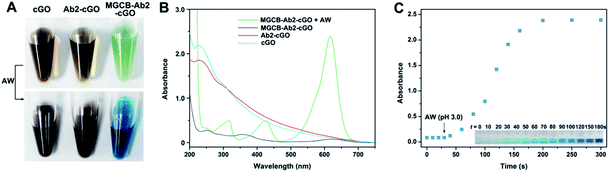
After obtaining the MGCB–Ab2–cGO complex, we demonstrate that MGCB molecules can effectively detach from cGO surfaces and lead to a color change in an acidic environment. Considering the poor solubility of MGCB in aqueous solution, we first compare the solubility of MGCB (10 μM) in water with different pH levels (Fig. S1†), and the results clearly show that it gradually becomes soluble and dark turquoise when the pH level is below 6.0. So the water with a pH of 3.0 has been subsequently used to release the MGCB molecules on the cGO surface. From Fig. 3A, it is observed that cGO and Ab2–cGO share the same yellowish brown color, while MGCB–Ab2–cGO turns bright green. After adding AW (pH 3.0) into the three solutions, only the color of the MGCB–Ab2–cGO complex changes to turquoise quickly. This phenomenon is attributed to the transformation of MGCB from a neutral hydrophobic moiety to a cationic hydrophilic moiety. The color change is accompanied by the appearance of new absorption peaks at 316, 425 and 617 nm in the UV-vis spectrum (Fig. 3B). Also of note, there is no color response in the supernatant after centrifugation of MGCB–Ab2–cGO, which demonstrates the negligible leak of MGCB. As shown in Fig. 3C, the color recovery process of the MGCB–Ab2–cGO complex in the presence of AW is time-dependent. The MGCB absorbance increases significantly over 3 min and then remains constant without fading. Additionally, we have also optimized the concentration of MGCB modified on the surface of cGO in this system. With the increase of MGCB in a fixed concentration of cGO (10 μg), the color recovers gradually and the maximum value is reached when the concentration of MGCB is increased up to 3 mM. Thus, considering the high surface area of graphene (the calculated value is 2630 m2 g−1, which includes both surfaces), the maximum coverage of MGCB on the cGO surface is estimated to be ∼1.67 × 1014/cm2 with the assistance of a standard curve of MGCB (Fig. S2†).
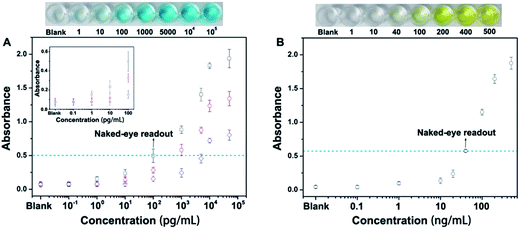
Next, we have employed this nanoprobe to develop a novel allochroic-cGO linked immunosorbent assay (ALISA) for protein detection that allows naked-eye signal readout with high sensitivity. We first use the MGCB-based ALISA to detect a model target, human IgG. In this case, Ab1 is mouse anti-human IgG monoclonal antibody, and Ab2 is goat anti-human IgG. The detection is performed in commonly used 96-well polystyrene (PS) plates, and the procedure for this assay is similar to conventional ELISA. Fig. 4A clearly shows that (1) higher concentrations of human IgG can capture more MGCB–Ab2–cGO. Upon the addition of AW (pH 3.0), more MGCB molecules can be released from the cGO surfaces, thus resulting in stronger absorbance recovery. (2) ALISA can detect human IgG as low as 1.0 pg mL−1. Also, the control experiment shows that the solution remains clear in the absence of the target, suggesting that it is the biospecific recognition between MGCB–Ab2–cGO and the target protein that results in the color change, rather than the nonspecific absorption of MGCB–Ab2–cGO on the PS surface. (3) The detection dynamic range can be conveniently controlled by tuning the loading capacity of MGCB on the cGO surface.
We have also compared the detection performance of ALISA with conventional ELISA that utilizes HRP-labeled antibody for enzyme-catalyzed signal transduction and amplification. As shown in Fig. 4A, ALISA allows a naked-eye readout when the concentration of human IgG is 100 pg mL−1. In comparison, a concentration of 40 ng mL−1 is required in conventional ELISA for a visible readout. This result represents a 400-fold increase in the sensitivity of the naked-eye detection (Fig. 4B). Also of note, conventional ELISA often takes 20 min or longer for final visualization and blocking buffer is essential to ensure accurate quantification, but only a 3 min incubation after adding AW is enough for ALISA, further simplifying the experimental procedure. Naked-eye detection is particularly useful in a variety of environments where access to laboratory equipment such as a plate reader is lacking. Therefore, developing simple and sensitive sensors with naked-eye readout may have great potential for improving the healthcare level of people in resource-poor regions.
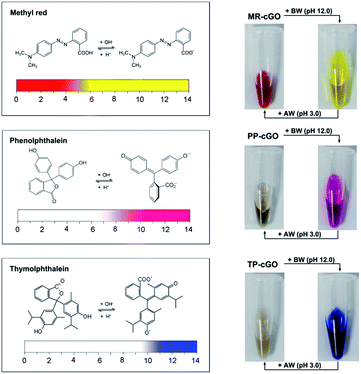
After demonstrating the analytical performance of our ALISA for a single target, we have further tested its multicolor detection ability which is hard to realize using conventional ELISA. Considering the limited number of color-changing organic bases, we have selected another three allochroic reagents (Fig. 5), methyl red (MR), phenolphthalein (PP) and thymolphthalein (TP). They are chosen as chromogenic agents due to the fact that (1) all of them are commonly-used acid–base indicators that present different colors such as yellow, pink, and blue when the pH level is above 10.0.21 (2) All of them contain two or three aromatic rings that are beneficial for π–π stacking interactions with graphene nanosheets. (3) All of them are easily discharged from the cGO surface after adding BW (pH 12.0). As anticipated, it is observed that three stable nanoprobes (MR–cGO, PP–cGO, and TP–cGO) can be easily obtained by mixing cGO with the three insoluble indicators, the maximum loading capacities of which are calculated to be 1.83 × 1014, 3.65 × 1013, and 8.9 × 1013/cm2 according to the standard curves (Fig. S3–S5†). Based on the above mentioned properties, it is easy to achieve detection performances similar to the MGCB-based ALISA.
As shown in Fig. S6,† the limit of detection (LOD) of the MR-, PP- and TP-based ALISA for IgG is determined to be 20, 1 and 0.1 pg mL−1 (Fig. S6–S8†), respectively. The difference in the LOD may be a result of the different load capacity and extinction coefficient of the three dyes.
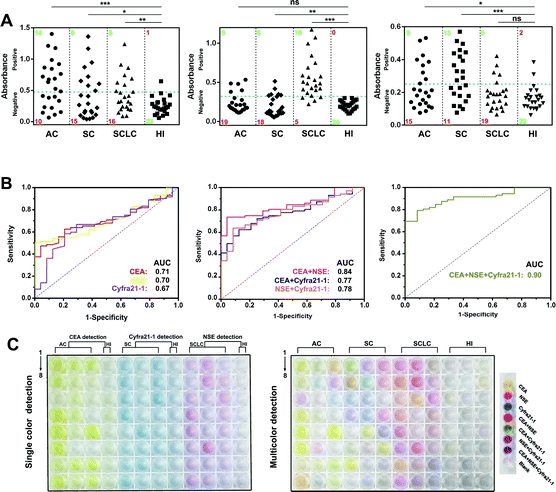
Then, we prepared MR–Ab2–cGO, PP–Ab2–cGO, and TP–Ab2–cGO to detect three tumor markers (TMs), including carcino-embryonic antigen (CEA), neuron-specific enolase (NSE), and cytokeratin-19 fragment (Cyfra 21-1). It was proposed that a high specificity for small-cell and non-small cell lung carcinoma (SCLC and NSCLC) in patient serum could be achieved by conjugating the nanoprobes with corresponding detection antibodies. Patient serum with SCLC or NSCLC, including adenocarcinoma (AC) and squamous carcinoma (SC), was collected from a local hospital (Table S1†). From the results shown in Fig. 6A (left panel), it can be determined that in the individual assay for CEA, the sensitivities for AC, SC, and SCLC are 58.3%, 37.5% and 33.3%, indicating its potential value for AC justification. Whereas NSE and Cyfra 21-1 display the highest sensitivities for SCLC (83.3%) and SC (54.2%) patients, respectively (Fig. 6A, middle and right panels). Although there are significant differences between the three TMs in a given lung cancer subtype and control group [healthy individual (HI), P < 0.001], it should be noted that any single one of them is not enough for the highly specific detection of lung cancer in patients (Table S4†). However, remarkable improvements in disease classification may be gained by using more than one biomarker. For example, the combination of CEA and NSE is found to be more effective in the diagnosis of lung cancer in patients with a sensitivity of 79.1%. When the detection of three TMs is combined together, it is found to be highly valuable in differentiating lung cancer patients with a sensitivity of 90.2% (Table S2†). To further characterize the discriminatory sensitivity and specificity of these TMs, we have further analyzed the biomarker responses in our ALISA using receiver operating characteristic (ROC) curves. The best individual biomarker for disease classification does not fully recapitulate the area under the curves (AUCs) ranging from 0.66 to 0.72 (Fig. 6B, left panel), but combinatorial panels, such as the best dual (NSE and CEA) and triple biomarker combinations, lead to improvements in predictivity (0.84 and 0.9, respectively; middle and right panels). In Fig. 6C, it can be seen that the final colors of the liquids correspond to collected patient samples. Notably, multicolor readout can be realized in a single run compared with the tedious one by one analysis of samples. Taken together, multicolor detection provides a more intuitive and convenient approach that facilitates the diagnosis of lung cancer in patients compared with single color detection.
Conclusions
In this study, we have fabricated a novel allochroic cGO-based nanoprobe to develop a multicolor immunoassay named as allochroic-cGO linked immunosorbent assay (ALISA), which allows multicolor detection of protein molecules at a pg mL−1 level. Not only is the LOD of our ALISA several orders of magnitude lower than for conventional HRP-based immunoassays, but this new assay method can also realize multicolor detection. The ultrahigh sensitivity of this nanoprobe is based on the high loading efficiency of allochroic dyes onto the cGO surface and high absorbance recovery, whereas the multicolor detection ability is derived from the release of different allochroic molecules from the cGO surface by simply adding AW or BW. Such improved detection systems would enable the rapid and sensitive screening of biomarkers in their early stages, thus providing improved diagnostic outcomes. In this work, the detection using our ALISA has also been performed in 96-well PS plates, the most popular detection format in clinical laboratories, especially in developing countries, making this method low-cost and easily adaptable into currently available diagnostic platforms.Acknowledgements
This work is supported by the National Natural Science Foundation of China (Grant No. 21235003, 21327902, J1210026), and the National Science Fund for Distinguished Young Scholars (Grant No. 20925520).Notes and references
- R. M. Lequin, Clin. Chem., 2005, 51, 2415 CAS.
- E. Engvall and P. Perlmann, Immunochemistry, 1971, 8, 871 CrossRef CAS PubMed.
- G. A. Kwong, G. von Maltzahn, G. Murugappan, O. Abudayyeh, S. Mo, I. A. Papayannopoulos, D. Y. Sverdlov, S. B. Liu, A. D. Warren, Y. Popov, D. Schuppan and S. N. Bhatia, Nat. Biotechnol., 2013, 31, 63 CrossRef CAS PubMed.
- G. F. Zheng, F. Patolsky, Y. Cui, W. U. Wang and C. M. Lieber, Nat. Biotechnol., 2005, 23, 1294 CrossRef CAS PubMed.
- Y. Xianyu, Z. Wang and X. Jiang, ACS Nano, 2014, 8, 12741 CrossRef CAS PubMed.
- A. P. Acharya, P. M. Nafisi, A. Gardner, J. L. MacKay, K. Kundu, S. Kumar and N. Murthy, Chem. Commun., 2013, 49, 10379 RSC.
- S. Tong, B. Ren, Z. Zheng, H. Shen and G. Bao, ACS Nano, 2013, 7, 5142 CrossRef CAS PubMed.
- S. I. Stoeva, J. S. Lee, J. E. Smith, S. T. Rosen and C. A. Mirkin, J. Am. Chem. Soc., 2006, 128, 8378 CrossRef CAS PubMed.
- M. Hu, J. Yan, Y. He, H. Lu, L. Weng, S. Song, C. Fan and L. Wang, ACS Nano, 2010, 4, 488 CrossRef CAS PubMed.
- R. S. Gaster, D. A. Hall, C. H. Nielsen, S. J. Osterfeld, H. Yu, K. E. Mach, R. J. Wilson, B. Murmann, J. C. Liao, S. S. Gambhir and S. X. Wang, Nat. Med., 2009, 15, 1327 CrossRef CAS PubMed.
- Y. Wan, Y.-G. Zhou, M. Poudineh, T. S. Safaei, R. M. Mohamadi, E. H. Sargent and S. O. Kelley, Angew. Chem., Int. Ed., 2014, 53, 13145 CrossRef CAS PubMed.
- Z. J. Zhang, Y. J. Guan, M. Li, A. D. Zhao, J. S. Ren and X. G. Qu, Chem. Sci., 2015, 6, 2822 RSC.
- R. de la Rica and M. M. Stevens, Nat. Nanotechnol., 2012, 7, 821–824 CrossRef CAS PubMed.
- D. Cecchin, R. de la Rica, R. E. S. Bain, M. W. Finnis, M. M. Stevens and G. Battaglia, Nanoscale, 2014, 6, 9559 RSC.
- M. Irie, J. Am. Chem. Soc., 1983, 105, 2078 CrossRef CAS.
- J. S. Lee, H.-A. Joung, M.-G. Kim and C. B. Park, ACS Nano, 2012, 6, 2978 CrossRef CAS PubMed.
- S. Song, Y. Xue, L. Feng, H. Elbatal, P. Wang, C. N. Moorefield, G. R. Newkome and L. Dai, Angew. Chem., Int. Ed., 2014, 53, 1415 CrossRef CAS PubMed.
- X. Sun, Z. Liu, K. Welsher, J. T. Robinson, A. Goodwin, S. Zaric and H. Dai, Nano Res., 2008, 1, 203 CrossRef CAS PubMed.
- W. Hu, G. He, H. Zhang, X. Wu, J. Li, Z. Zhao, Y. Qiao, Z. Lu, Y. Liu and C. M. Li, Anal. Chem., 2014, 86, 4488 CrossRef CAS PubMed.
- G. W. Stewart, Phys. Rev., 1929, 33, 0889 CrossRef CAS.
- P. Atkins, Chemical Principles, W. H. Freeman and Company, New York, 5th edn, 2009, p. 492 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5sc04256a |
| This journal is © The Royal Society of Chemistry 2016 |