Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Mucin architecture behind the immune response: design, evaluation and conformational analysis of an antitumor vaccine derived from an unnatural MUC1 fragment†
Nuria
Martínez-Sáez
a,
Nitin T.
Supekar
b,
Margreet A.
Wolfert
b,
Iris A.
Bermejo
a,
Ramón
Hurtado-Guerrero
cd,
Juan L.
Asensio
e,
Jesús
Jiménez-Barbero
fgh,
Jesús H.
Busto
a,
Alberto
Avenoza
a,
Geert-Jan
Boons
*b,
Jesús M.
Peregrina
*a and
Francisco
Corzana
*a
aDepartamento de Química, Universidad de La Rioja, Centro de Investigación en Síntesis Química, Madre de Dios 53, 26006 Logroño, Spain. E-mail: francisco.corzana@unirioja.es; jesusmanuel.peregrina@unirioja.es
bComplex Carbohydrate Research Center, University of Georgia, 315 Riverbend Road, Athens, Georgia 30602, USA. E-mail: gjboons@ccrc.uga.edu
cBIFI, University of Zaragoza, BIFI-IQFR (CSIC) Joint Unit, Mariano Esquillor s/n, Campus Rio Ebro, Edificio I+D, Zaragoza, Spain
dFundación ARAID, 50018, Zaragoza, Spain
eInstituto de Química Orgánica General, IQOG-CSIC, Juan de la Cierva 3, 28006 Madrid, Spain
fStructural Biology Unit, CIC bioGUNE, Parque Tecnológico de Bizkaia Building 801A, 48160 Derio, Spain
gIKERBASQUE, Basque Foundation for Science, 48011 Bilbao, Spain
hDepartment of Chemical and Physical Biology, Centro de Investigaciones Biológicas, CSIC, Ramiro de Maeztu 9, 28040 Madrid, Spain
First published on 15th December 2015
Abstract
A tripartite cancer vaccine candidate, containing a quaternary amino acid (α-methylserine) in the most immunogenic domain of MUC1, has been synthesized and examined for antigenic properties in transgenic mice. The vaccine which is glycosylated with GalNAc at the unnatural amino acid, was capable of eliciting potent antibody responses recognizing both glycosylated and unglycosylated tumour-associated MUC1 peptides and native MUC1 antigen present on cancer cells. The peptide backbone of the novel vaccine presents the bioactive conformation in solution and is more resistant to enzymatic degradation than the natural counter part. In spite of these features, the immune response elicited by the unnatural vaccine was not improved compared to a vaccine candidate containing natural threonine. These observations were rationalized by conformational studies, indicating that the presentation and dynamics of the sugar moiety displayed by the MUC1 derivative play a critical role in immune recognition. It is clear that engineered MUC1-based vaccines bearing unnatural amino acids have to be able to emulate the conformational properties of the glycosidic linkage between the GalNAc and the threonine residues. The results described here will be helpful to the rational design of efficacious cancer vaccines.
Introduction
Mucin MUC1 is an O-glycoprotein strongly over-expressed in various tumours.1–5 In healthy tissues, the MUC1 backbone presents complex oligosaccharides, while in cancer cells it shows simple and truncated carbohydrates because of the incomplete glycosylation. As a result, different tumour-associated carbohydrate antigens (TACAs), such as the Tn determinant (α-O-GalNAc-Ser/Thr),6 are exposed to the immune system and can be recognized by different antibodies.7 Due to this unique characteristic, MUC1 derivatives are attracting great interest as a potential tool in developing therapeutic vaccines for the treatment of cancer.3,8–10 However, to date, none of them have succeeded in clinical trials11 due to the fact that most natural TACAs are tolerated by the immune system and glycosidases can reduce their in vivo bioavailability.12 One way to overcome this issue is through the use of chemical modifications of the antigens to generate non-natural determinants.13 A number of TACA mimics, comprising C-glycosides14–16 and S-glycosides,17–19 have been incorporated into carbohydrate-based vaccines. Additionally, the use of homoserine and β3-homothreonine conjugates20,21 to construct mucin-like glycopeptides and derivatives that incorporate fluorine atoms22,23 have been proposed.Nevertheless, it is important to mention that the structural basis for the design of these vaccines remain unclear. From the view of molecular recognition, the Pro-Asp-Thr-Arg (PDTR) sequence comprises the minimal epitope for most of anti-MUC1 antibodies,7 such as SM3,24 which has a potential use in the early diagnosis and treatment of breast cancer.25 Although the crystal structure of this antibody in complex with a small peptide was reported some years ago,25 we have recently reported the X-ray structures of short glycopeptides bound to SM3.26 The analysis of these structures reveals that the threonine (Thr) residue of this epitope adopts a helix-like conformation and that its β-methyl is engaged in a hydrophobic contact with the surface of the antibody.
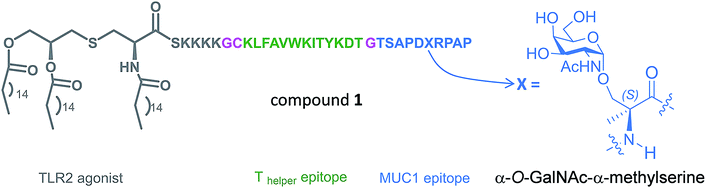
With these considerations in mind, we designed a novel Tn antigen mimic based on the quaternary amino acid α-methylserine (MeSer).27 This amino acid favors helix-like structures28 and has a methyl group at Cα that can establish the hydrophobic contact commented above with the antibody.
In the present work, we have incorporated this unnatural Tn into the most immunogenic domain of a MUC1 fragment, designing a three-component cancer vaccine (Fig. 1), similar to that previously reported by Boons and co-workers.8 We have demonstrated that the unnatural glycosylated epitope exhibits better stability in human serum when compared to the natural derivative. The novel vaccine is able to elicit a potent immune response in transgenic mice, recognizing both glycosylated and unglycosylated tumour-associated MUC1 derivatives and native MUC1 antigen present on cancer cells. The effectiveness of this vaccine is comparable (but not better) to that observed for its homologue derivative with threonine. To explain the experimental data, we have performed an extensive conformational analysis on the vaccine in the free state in water as well as bound to phospholipid-based liposomes. The analysis involves the use of NMR and Molecular Dynamics (MD) simulations. The conformational studies point out that the extra flexibility of the side chain and the glycosidic linkage of the unnatural GalNAc-MeSer fragment have an unfavorable impact on the molecular recognition by the immune system. This information reinforces the idea that the design of more efficient vaccines based on MUC1 has to involve the use of glycopeptides that can imitate the conformational preferences of the aberrantly glycosylated natural MUC1 epitope.
Results and discussion
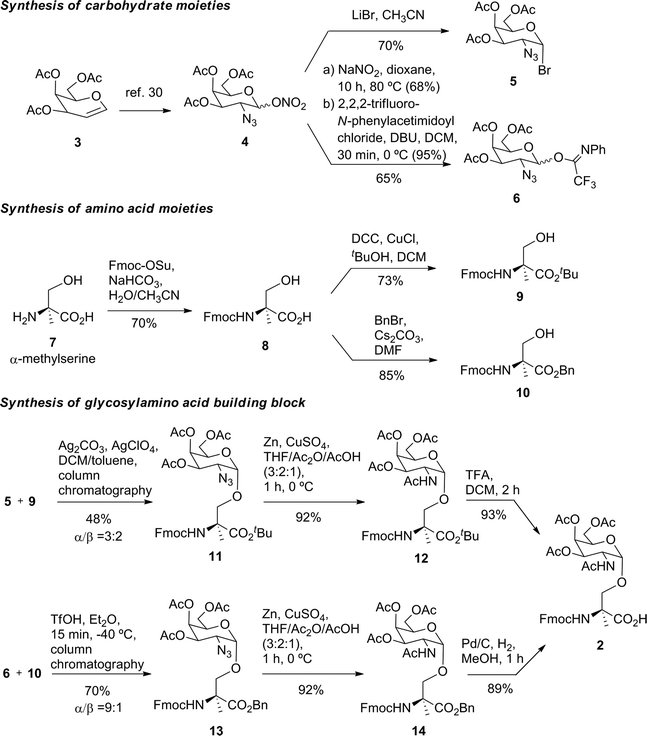
Prior to the preparation of vaccine candidate 1, we optimized the synthesis of the unnatural building block 2 (Scheme 1) in a large scale. To this end, we developed two divergent synthetic routes that are summarized in Scheme 1. The first one uses derivative 5 as a glycosyl donor, which was easily prepared from compound 4 and following the methodology described in the literature.29 Derivative 4 was obtained from commercially available 3,4,6-tri-O-acetyl-D-galactal 3 (see ref. 30). The adequately protected α-methylserine 9, as N-Fmoc31 and tert-butyl ester32 protecting groups, was synthesized starting from α-methylserine 7, obtained by methodology described by us.33 Initially, to carry out the glycosylation of protected amino acid 9 with derivative 5, we examined the method of Liebe and Kunz34 using AgCO3 and AgClO4 in a mixture of toluene and dichloromethane (DCM), obtaining a mixture of α/β anomers in a ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 in a moderate yield. The α-anomer 11 was purified by column chromatography and its azide group was transformed into an acetamido group by reduction with Zn in the presence of CuSO4 to give compound 12 in an excellent yield. The tert-butyl ester group of this compound was removed to afford the required building block 2. In order to improve the yield of the glycosylation reaction, as well as the anomeric ratio, we used a modification of the trichloroacetimidate method.35 Therefore, the imidate-derivative 6 was prepared by hydrolysis of the nitro group of compound 4 (ref. 36), followed by treatment with 2,2,2-trifluoro-N-phenylacetimidoyl chloride in the presence of DBU. The use of the acid glycosylation method forced us to prepare a different and convenient protected quaternary amino acid. N-Fmoc-α-methylserine 8 was converted into benzyl ester derivative 10 using benzyl bromide in the presence of cesium carbonate. The glycosylation of this compound with imidate 6 was carried out with triflic acid in diethyl ether at −40 °C for 15 min, giving a 70% yield of α/β anomers in a ratio 9
2 in a moderate yield. The α-anomer 11 was purified by column chromatography and its azide group was transformed into an acetamido group by reduction with Zn in the presence of CuSO4 to give compound 12 in an excellent yield. The tert-butyl ester group of this compound was removed to afford the required building block 2. In order to improve the yield of the glycosylation reaction, as well as the anomeric ratio, we used a modification of the trichloroacetimidate method.35 Therefore, the imidate-derivative 6 was prepared by hydrolysis of the nitro group of compound 4 (ref. 36), followed by treatment with 2,2,2-trifluoro-N-phenylacetimidoyl chloride in the presence of DBU. The use of the acid glycosylation method forced us to prepare a different and convenient protected quaternary amino acid. N-Fmoc-α-methylserine 8 was converted into benzyl ester derivative 10 using benzyl bromide in the presence of cesium carbonate. The glycosylation of this compound with imidate 6 was carried out with triflic acid in diethyl ether at −40 °C for 15 min, giving a 70% yield of α/β anomers in a ratio 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. α-Anomer 13 was purified by silica gel column chromatography and its azide group was reduced to acetamido group to give compound 14.
1. α-Anomer 13 was purified by silica gel column chromatography and its azide group was reduced to acetamido group to give compound 14.
The benzyl ester of the latter compound was removed by hydrogenolysis, giving building block 2 in a good overall yield (Scheme 1).
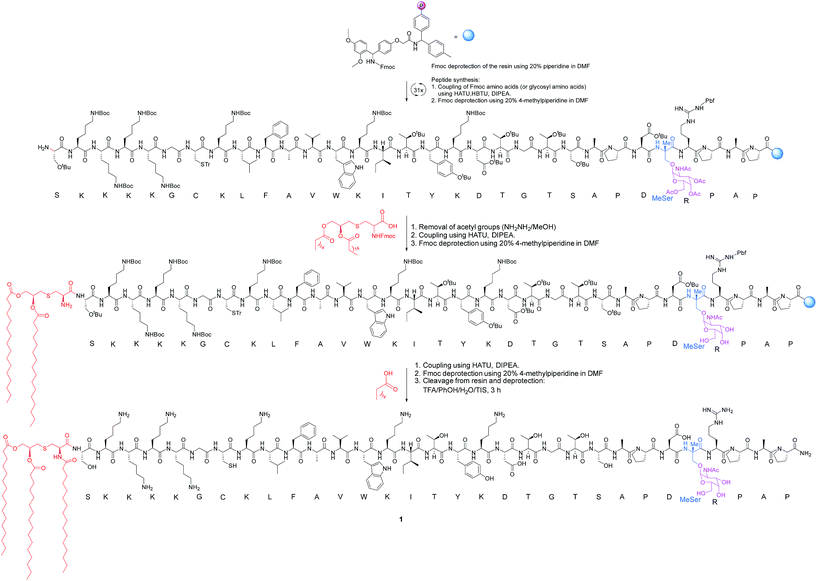
The synthesis of vaccine candidate 1 was carried out by microwave-assisted solid phase peptide synthesis (MW-SPPS) employing a Rink amide AM LL resin, Fmoc-protected amino acids and derivative 2 (Scheme 2).35 The incorporation of building block 2 and the assembly of Fmoc-Pam2-Cys-OH were manually performed under microwave-assisted coupling conditions.
We examined then the stability of the unnatural MUC1 derivative in human plasma. To this end, we synthesized glycopeptides APDThr(α-D-GalNAc)RP and APDMeSer(α-D-GalNAc)RP (ESI†). The degradation of the glycopeptide containing threonine was faster (around 1.8-fold) compared to the unnatural compound. We hypothesized that this resistance may translate to an increase in in vivo stability and bioavailability and hence lead to stronger and longer-lasting antigenic responses.
To examine the immunity of lipoglycopeptide 1, it was incorporated into phospholipid-based small vesicles (ESI†). The aliphatic chain of the palmitic acid in the TLR2 agonist favours the incorporation of the vaccine into liposomes, which may enhance the circulation time of the vaccine and allows for the presentation of the glycopeptide epitope in a multivalent manner.8
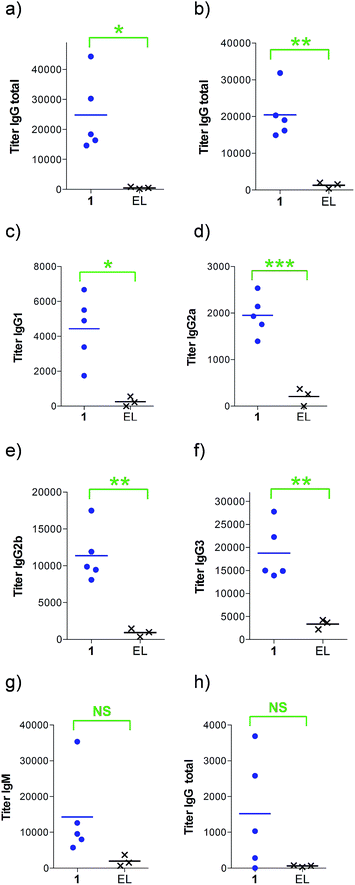
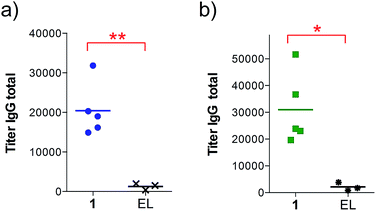
Next, groups of MUC1.Tg mice (C57BL/6; H-2b) that express human MUC1 (ref. 37) were immunized four times at biweekly intervals with liposomal preparations of compound 1 and empty liposomes as control. One week after the last immunization, the mice were sacrificed and the serum harvested. Mice immunized with 1 elicited a specific anti-MUC1 IgG antibody response (Table 1 and Fig. 2), which was significantly different from the control group immunized with empty liposomes (EL). Sub-typing of the IgG antibodies shows significant IgG1 and IgG2a,b responses, indicating a mixed Th1/Th2 response.
| Immunizationb | 1 | ELd |
|---|---|---|
| a Anti-MUC1 and anti-Thelper antibody titers are presented as median values for groups of mice. ELISA plates were coated with BSA-MI-CTSAPDT(α-D-GalNAc)RPAP conjugate for anti-MUC1 antibody titers or NeutrAvidin-biotin-Thelper for anti-Thelper antibody titers. Titers were determined by linear regression analysis, with plotting of dilution versus absorbance. Titers are defined as the highest dilution yielding an optical density of 0.1 or greater relative to normal control mouse sera. b Liposomal preparations were employed. c EP stands for endpoint (serum samples after 4 immunizations). d EL stands for empty liposomes. MI stands for maleimide. Individual anti-MUC1 titers for IgG total, IgG1, IgG2a, IgG2b, IgG3 and IgM, and anti-Thelper epitope for IgG total are reported in Fig. 2. | ||
| IgG total, MUC1, 3rd imm | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
800 |
| IgG total, MUC1, EPc | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
1500 |
| IgG1, MUC1, EP | 4900 | 600 |
| IgG2a, MUC1, EP | 1900 | 0 |
| IgG2b, MUC1, EP | 9900 | 1000 |
| IgG3, MUC1, EP | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
400 |
| IgM, MUC1, EP | 9600 | 100 |
| IgG total, Thelper, EP | 1000 | 100 |
The observed high IgG3 titer is typical of an anti-carbohydrate response. IgM antibody titers were low and not significantly different from the control. Low amounts of IgG antibodies raised against the Thelper epitope were measured; indicating that candidate vaccine 1 does not suffer from significant immune suppression.
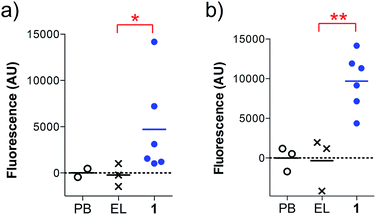
We found that the antibodies produced by vaccine 1 could recognize glycosylated and unglycosylated MUC1 epitopes with similar titers (Fig. 3), indicating that antibodies recognize the peptide and not the glycan.
In addition, the ability of the mouse antisera to recognize the MUC1 antigen expressed on cancer cells was investigated. Serum samples were added to MUC1 expressing MCF7 human breast cancer and C57mg.MUC1 mammary gland tumor cells, and recognition was established using a FITC-labeled anti-mouse IgG antibody. As shown in Fig. 4, antisera obtained from immunizations with vaccine 1 displayed recognition of MUC1 tumour cells, whereas no binding was observed for the controls.
Of note, vaccine 1 did not improve the effectiveness of a previously reported natural threonine-containing derivative8 (ESI†). Prompted by these results, we investigated whether the unnatural scaffold influences the presentation of the epitope. To this end, we ran 100 ns MD simulations on phospholipid-based liposomes loaded with only a lipoglycopeptide 1 molecule to simplify the system. For comparative purposes, we performed also the MD simulation on the threonine analogue. According to these simulations, which start with an extended conformation of the peptidic backbone, both lipoglycopeptides have a clear tendency to adopt folded conformations, allowing the sugar moiety to be close to the liposome surface at the end of the MD simulations. This feature is more marked in the analogue with threonine.
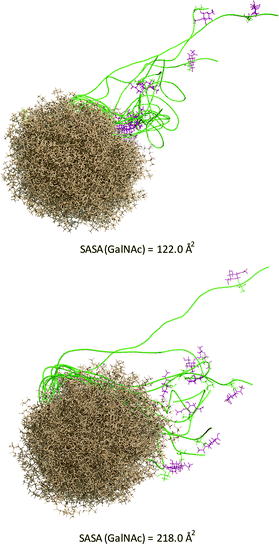
The solvent-accessible surface area (SASA) calculated for the GalNAc residue, which gives an idea of its accessibility, is ca. 1.8-fold higher for the unnatural vaccine (Fig. 5), indicating that the unnatural scaffold does not hinder the Tn-mimicry to interact with the immune system.
Attending to our simulations, this result does not explain the experimental evidence. Accordingly, we focused our conformational analysis on the epitope fragments. Thus, glycopeptides g1 and g2 (Fig. 6a) were synthesized and subjected to detailed conformational analysis by combining NMR and MD simulations. The proton–proton distances obtained from 2D-NOESY experiments (Fig. 6a) were used as restraints in MD simulations (MD-tar).38,39 This type of calculation provides a distribution of low energy conformers able to reproduce the experimental data. The good agreement obtained in our simulations between the experimental and calculated distances validates the theoretical study (see ESI†).
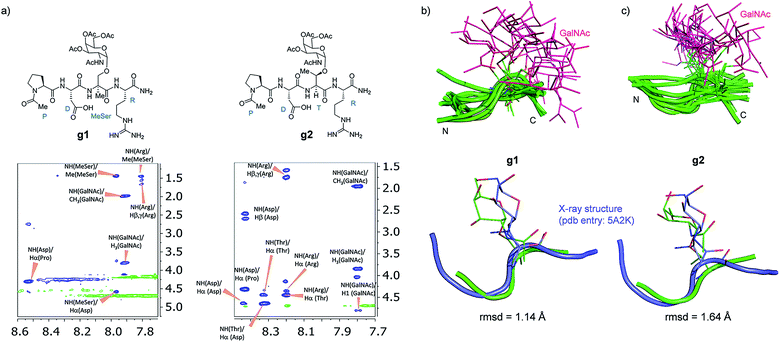 | ||
Fig. 6 (a) Glycopeptides g1 and g2 and sections of the 500 ms 2D NOESY spectrum (400 MHz) in H2O/D2O (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) at 25 °C and pH = 5.6 showing the amide cross-peaks. Diagonal peaks and exchange cross-peaks connecting NH protons and water are negative (green color). The NOE contacts are represented as positive cross-peaks (blue color). Ensembles obtained for glycopeptides g1 (b) and g2 (c) through the 20 ns MD-tar simulations in water, together with the main conformation found in solution for the backbone of the glycopeptides (lower panels). The backbone of g1 and g2 are superimposed onto the geometry of the APDThr(α-O-GalNAc)RP glycopeptide in complex with SM3 antibody (PDB entry: 5A2K). rmsd = root-mean-square deviation. 1) at 25 °C and pH = 5.6 showing the amide cross-peaks. Diagonal peaks and exchange cross-peaks connecting NH protons and water are negative (green color). The NOE contacts are represented as positive cross-peaks (blue color). Ensembles obtained for glycopeptides g1 (b) and g2 (c) through the 20 ns MD-tar simulations in water, together with the main conformation found in solution for the backbone of the glycopeptides (lower panels). The backbone of g1 and g2 are superimposed onto the geometry of the APDThr(α-O-GalNAc)RP glycopeptide in complex with SM3 antibody (PDB entry: 5A2K). rmsd = root-mean-square deviation. | ||
Fig. 6b and c show the ensemble obtained from 20 ns MD-tar simulations in explicit water. The unnatural residue in compound g1 adopts a helix-like conformation in water, which is validated by the NOE cross-peaks between the methyl group of MeSer and NH protons of MeSer and Arg residues (Fig. 6a). The folded conformation adopted by the unnatural residue is in agreement with previous studies on other glycopeptides carrying this quaternary amino acid.27 However, the presence of α-methyl group provides flexibility to the peptide backbone. The most populated conformer (30%) is similar to the bioactive conformation found in the crystal structures.26 This conformation is barely populated in the free state for glycopeptide g2, which exhibits an extended conformation (around 78%) for the backbone, in agreement with previous structural studies on MUC1-derived glycopeptides modified by GalNAc.26,40–43 Concerning the glycosidic linkage, it adopts the ‘alternate’ conformation in the unnatural glycopeptide g1, with ϕ torsional angle around 80°, in agreement with the exo-anomeric effect, and with the ψ dihedral close to 180° (Fig. 7). This geometry of the glycosidic linkage is, as expected,44 observed in the analogue glycopeptide with serine also studied in this work (ESI†). In contrast, the ‘eclipsed’ conformation, with ϕ around 120°, is preferred in glycopeptide g2 (ref. 45). This particular conformation is characterized by a NOE cross-peak between the amide groups of GalNAc and Thr (ESI†). Consequently, the carbohydrate moiety adopts different orientations with respect to the peptide backbone in the investigated compounds. Regarding the side chain of the glycosylated residue, while in g2 it is locked, showing a value close to 60°, in glycopeptide g1 the χ1 torsional angle is rather flexible, showing two similar populated conformers, with values around 60° [g(+)] and 180° [anti] (Fig. 7). These conformational differences were also observed when peptide fragments, g1 (amino acids in blue in Fig. 1) and g2 (natural epitope), are part of the 10-mer MUC1 derivatives, indicating the generality of the different conformational behavior. These findings were deduced from the 100 ns MD simulations carried out on the liposomes (Fig. 5 and ESI†). Therefore, although the unnatural glycopeptide exhibits a higher population of the bioactive backbone conformation with respect to the natural epitope, it displays a higher flexibility at the sugar moiety. Presumably, the extra flexibility of the unnatural epitope will have a negative impact on the molecular recognition by the immune system. This hypothesis might be validated by the excellent results recently reported by Nativi and co-workers with a non-natural vaccine based on a conformational restricted Tn antigen.13 In summary, our data reinforces the idea that the design of more efficient cancer vaccines has to involve the use of glycopeptides that can emulate the peculiar conformational preferences of the α-GalNAc-Thr glycosidic linkage,45 found in aberrantly glycosylated natural MUC1 epitope.
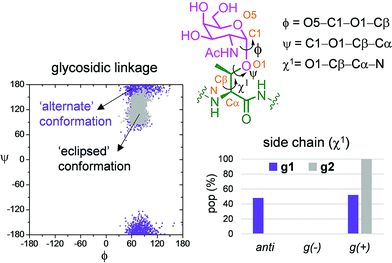 | ||
| Fig. 7 Differences obtained from the MD-tar simulations between the glycosidic linkage and the side chain of α-methylserine and threonine residues in compounds g1 and g2, respectively. | ||
Conclusions
A MUC1-based vaccine containing the quaternary amino acid α-methylserine at the most immunogenic domain has been synthesized and the immunogenicity examined in transgenic mice. The vaccine elicits robust antibody titers recognizing glycosylated and unglycosylated tumour-associated MUC1 derivatives and native MUC1 antigen present on cancer cells. In spite of the peptide backbone of the novel vaccine presenting the bioactive conformation in solution and it being more resistant to enzymatic degradation than the natural one, the immune response elicited by this unnatural vaccine is not superior to the threonine analogue. To rationalize these observations, we have performed conformational studies on the unnatural vaccine and on its natural counterpart. These studies indicate that the presentation and dynamics of the sugar moiety displayed by the MUC1 derivative may be critical for a strong immune response. The present work offers a new way to engineer MUC1-based vaccines based on the conformational behavior of their components. The designed unnatural amino acids have to be able to emulate the particularities of the glycosidic linkage between the GalNAc and the threonine residues. Our results provide foundational information for the rational design of carbohydrate-based vaccines against cancer.Acknowledgements
We thank the Ministerio de Economía y Competitividad (projects CTQ2012-36365/FEDER, CTQ2012-32025/FEDER, CTQ2013-44367-C2-2-P, UNLR13-4E-1931). N. M.-S. thanks the Universidad de La Rioja for the FPI grant. I. A. B. thanks the Asociación Española Contra el Cáncer en La Rioja for a grant. We also thank CESGA for computer support. G.-J. B. acknowledges the National Cancer Institute of the US National Institutes of Health for support (R01CA88986). Animal studies have been approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Georgia.Notes and references
- N. Gaidzik, U. Westerlind and H. Kunz, Chem. Soc. Rev., 2013, 42, 4421 RSC; C. Nativi and O. Renaudet, ACS Med. Chem. Lett., 2014, 5, 1176 CrossRef CAS PubMed.
- T. Buskas, P. Thompson and G.-J. Boons, Chem. Commun., 2009, 5335 RSC.
- R. M. Wilson and S. J. Danishefsky, J. Am. Chem. Soc., 2013, 135, 14462 CrossRef CAS PubMed; R. Adamo, A. Nilo, B. Castagner, O. Boutureira, F. Bertia and G. J. L. Bernardes, Chem. Sci., 2013, 4, 2995 RSC.
- D. W. Kufe, Nat. Rev. Cancer, 2009, 9, 874 CrossRef CAS PubMed.
- M. A. Tarp and H. Clausen, Biochim. Biophys. Acta, 2008, 1780, 546 CrossRef CAS PubMed.
- T. Ju, V. I. Otto and R. D. Cummings, Angew. Chem., Int. Ed., 2011, 50, 1770 CrossRef CAS PubMed.
- U. Karsten, N. Serttas, H. Paulsen, A. Danielczyk and S. Goletz, Glycobiology, 2004, 14, 681 CrossRef CAS PubMed.
- V. Lakshminarayanan, P. Thompson, M. A. Wolfert, T. Buskas, J. M. Bradley, L. B. Pathangey, C. S. Madsen, P. A. Cohen, S. J. Gendler and G.-J. Boons, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 261 CrossRef CAS PubMed.
- H. Cai, M.-S. Chen, Z.-Y. Sun, Y.-F. Zhao, H. Kunz and Y.-M. Li, Angew. Chem., Int. Ed., 2013, 52, 6106 CrossRef CAS PubMed.
- Z. Yin and X. Huang, J. Carbohydr. Chem., 2012, 31, 143 CrossRef CAS PubMed.
- D. Miles, H. RochØ, M. Martin, T. J. Perren, D. A. Cameron, J. Glaspy, D. Dodwell, J. Parker, J. Mayordomo, A. Tres, J. L. Murray and N. K. Ibrahim, Oncologist, 2011, 16, 1092 CrossRef CAS PubMed.
- C. Ohyama, Int. J. Clin. Oncol., 2008, 13, 308 CrossRef CAS PubMed.
- B. Richichi, B. Thomas, M. Fiore, R. Bosco, H. Qureshi, C. Nativi, O. Renaudet and L. BenMohamed, Angew. Chem., Int. Ed., 2014, 53, 11917 CrossRef CAS PubMed.
- A. Dondoni and A. Marra, Chem. Rev., 2000, 100, 4395 CrossRef CAS PubMed.
- B. Kuberan, S. A. Sikkander, H. Tomiyama and R. J. Linhardt, Angew. Chem., Int. Ed., 2003, 42, 2073 CrossRef CAS PubMed.
- L. Awad, R. Madani, A. Gillig, M. Kolympadi, M. Philgren, A. Muhs, C. Gérard and P. Vogel, Chem.–Eur. J., 2012, 18, 8578 CrossRef CAS PubMed.
- X. Wu, T. Lipinski, E. Paszkiewicz and D. R. Bundle, Chem.–Eur. J., 2008, 14, 6474 CrossRef CAS PubMed.
- D. R. Bundle, J. R. Rich, S. Jacques, H. N. Yu, M. Nitz and C.-C. Ling, Angew. Chem., Int. Ed., 2005, 44, 7725 CrossRef CAS PubMed.
- C.-X. Huo, X.-J. Zheng, A. Xiao, C.-C. Liu, S. Sun, Z. Lv and X.-S. Ye, Org. Biomol. Chem., 2015, 13, 3677 CAS.
- S. Vichier-Guerre, R. Lo-Man, V. Huteau, E. Dériaud, C. Leclerc and S. Bay, Bioorg. Med. Chem. Lett., 2004, 14, 3567 CrossRef CAS PubMed.
- A. S. Norgren and P. I. Arvidsson, Org. Biomol. Chem., 2005, 3, 1359 CAS.
- T. Oberbillig, C. Mersch, S. Wagner and A. Hoffmann-Röder, Chem. Commun., 2012, 48, 1487 RSC.
- M. Johannes, M. Reindl, B. Gerlitzki, E. Schmitt and A. Hoffmann-Röder, Beilstein J. Org. Chem., 2015, 11, 155 CrossRef PubMed.
- J. Burchell, S. Gendler, J. Taylor-Papadimitriou, A. Girling, A. Lewis, R. Millis and D. Lamport, Cancer Res., 1987, 47, 5476 CAS.
- P. Dokurno, P. A. Bates, H. A. Band, L. M. Stewart, J. M. Lally, J. M. Burchell, J. Taylor-Papadimitriou, D. Snary, M. J. Stern- Berg and P. S. Freemont, J. Mol. Biol., 1998, 284, 713 CrossRef CAS PubMed.
- N. Martínez-Sáez, J. Castro-López, J. Valero-González, D. Madariaga, I. Compañón, V. J. Somovilla, M. Salvadó, J. L. Asensio, J. Jiménez-Barbero, A. Avenoza, J. H. Busto, G. J. L. Bernardes, J. M. Peregrina, R. Hurtado-Guerrero and F. Corzana, Angew. Chem., Int. Ed., 2015, 54, 9830 CrossRef PubMed.
- F. Corzana, J. H. Busto, F. Marcelo, M. García de Luis, J. L. Asensio, S. Martín-Santamaría, Y. Sáenz, C. Torres, J. Jiménez-Barbero, A. Avenoza and J. M. Peregrina, Chem. Commun., 2011, 47, 5319 RSC.
- F. Corzana, J. H. Busto, F. Marcelo, M. García de Luis, J. L. Asensio, S. Martín-Santamaría, J. Jiménez-Barbero, A. Avenoza and J. M. Peregrina, Chem.–Eur. J., 2011, 17, 3105 CrossRef CAS PubMed.
- M. Liu, V. G. Young Jr, S. Lohani, D. Live and G. Barany, Carbohydr. Res., 2005, 340, 1273 CrossRef CAS PubMed.
- R. U. Lemieux and R. M. Ratcliffe, Can. J. Chem., 1979, 57, 1244–1251 CrossRef CAS; C. Heggemann, C. Budke, B. Schomburg, Z. Majer, M. Wißbrock, T. Koop and N. Sewald, Amino Acids, 2010, 38, 213 CrossRef PubMed.
- A. Paquet, Can. J. Chem., 1982, 60, 976–980 CrossRef CAS; G. F. Sigler, W. D. Fuller, N. C. Chaturvedi, M. Goodman and M. Verlander, Biopolymers, 1983, 22, 2157 CrossRef.
- M. Schultz and H. Kunz, Tetrahedron: Asymmetry, 1993, 4, 1205 CrossRef CAS.
- C. Aydillo, G. Jiménez-Osés, J. H. Busto, J. M. Peregrina, M. M. Zurbano and A. Avenoza, Chem.–Eur. J., 2007, 13, 4840 CrossRef CAS PubMed.
- B. Liebe and H. Kunz, Angew. Chem., Int. Ed., 1997, 36, 618 CrossRef CAS.
- R. C. Payne, S. Ficht, S. Tang, A. Brik, Y.-Y. Yang, D. A. Case and C. H. Wong, J. Am. Chem. Soc., 2007, 129, 13527 CrossRef CAS PubMed; P. Thompson, V. Lakshminarayanan, N. T. Supekar, J. M. Bradley, P. A. Cohen, M. A. Wolfert, S. J. Gendler and G.-J. Boons, Chem. Commun., 2015, 10214 RSC.
- K. Briner and A. Vasella, Helv. Chim. Acta, 1987, 70, 1341 CrossRef CAS.
- P. Mukherjee, C. S. Madsen, A. R. Ginardi, T. L. Tinder, F. Jacobs, J. Parker, B. Agrawal, B. M. Longenecker and S. J. Gendler, J. Immunother., 2003, 26, 47 CrossRef CAS PubMed.
- D. A. Pearlman and P. A. Kollman, J. Mol. Biol., 1991, 220, 457 CrossRef CAS PubMed.
- F. Corzana, J. H. Busto, S. B. Engelsen, J. Jiménez-Barbero, J. L. Asensio, J. M. Peregrina and A. Avenoza, Chem.–Eur. J., 2006, 12, 7864 CrossRef CAS PubMed.
- T. Matsushita, N. Ohyabu, N. Fujitani, K. Naruchi, H. Shimizu, H. Hinou and S.-I. Nishimura, Biochemistry, 2013, 52, 402 CrossRef CAS PubMed.
- S. Dziadek, C. Griesinger, H. Kunz and U. M. Reinscheid, Chem.–Eur. J., 2006, 12, 4981 CrossRef CAS PubMed.
- L. Kinarsky, G. Suryanarayanan, O. Prakash, H. Paulsen, H. Clausen, F.-G. Hanisch, M. A. Hollingsworth and S. Sherman, Glycobiology, 2003, 13, 929 CrossRef CAS PubMed.
- J. Schuman, A. P. Campbell, R. R. Koganty and B. M. Longenecker, J. Pept. Res., 2003, 61, 91 CrossRef CAS PubMed.
- F. Corzana, J. H. Busto, G. Jiménez-Osés, J. L. Asensio, J. Jiménez-Barbero, J. M. Peregrina and A. Avenoza, J. Am. Chem. Soc., 2006, 128, 14640 CrossRef CAS PubMed.
- F. Corzana, J. H. Busto, G. Jiménez-Osés, M. García de Luis, J. L. Asensio, J. Jiménez-Barbero, J. M. Peregrina and A. Avenoza, J. Am. Chem. Soc., 2007, 129, 9458 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Characterization of the new compounds, biological assays and MD simulations. See DOI: 10.1039/c5sc04039f |
| This journal is © The Royal Society of Chemistry 2016 |