PEGylated cationic hybrid bicellar nanodisc for efficient siRNA delivery†
Yanyan Li‡
a,
Yidi Wu‡c,
Shuquan Zhengc,
Xiaolong Liange,
Xiaorui Hanc,
Renfa Liud,
Deyao Zhaoc,
Yunhui Zhaof,
Yushen Jind,
Min Chend,
Xiaoxia Wangc,
Huiqing Caoc,
Xiuli Yue*b,
Tiejun Sten Shi*ag and
Zicai Liang*c
aSchool of Life Science and Technology, Harbin Institute of Technology, Harbin 150001, PR China
bSchool of Municipal and Environmental Engineering, Harbin Institute of Technology, Harbin 150001, PR China. E-mail: xiulidx@163.com
cLaboratory of Nucleic Acid Technology, Institute of Molecular Medicine, Peking University, Beijing 100871, PR China. E-mail: liangz@pku.edu.cn
dDepartment of Biomedical Engineering, College of Engineering, Peking University, Beijing 100871, PR China
eDepartment of Ultrasonography, Peking University Third Hospital, Beijing 100191, PR China
fSchool of Chemistry and Chemical Engineering, Hunan University of Science and Technology, Xiangtan 411201, PR China
gDepartment of Biomedicine, University of Bergen, Bergen, Norway. E-mail: tiejun.shi@uib.no
First published on 16th November 2016
Abstract
A hybrid cationic bicellar nanodisc was prepared by a conventional Bangham method in combination with a sol–gel reaction and self-assembly process. The relatively small size and disc-like shape was very stable because the incorporation of the organic–inorganic hybrid lipid had formed a crosslinked siloxane net structure on the surface. Physicochemical properties, biocompatibility, intracellular trafficking behavior, in vitro transfection efficiency and in vivo distribution of the bicellar nanodiscs were investigated. The unique characteristics of the cationic PEGylated hybrid bicellar nanodiscs address many of the deficiencies relating to current liposome technology. By optimizing the doping ratio of PEG-containing lipid, we found that a doping ratio of 1 mol% is enough to confer an excellent in vivo delivery performance while not compromising the transfection efficacy in vitro. The distinct disc-like shape, high stability conferred by the hybrid lipid and modest siRNA delivery performance make this platform promising as a siRNA vehicle for efficient siRNA delivery both in vitro and in vivo.
Introduction
RNA interference (RNAi) technology is emerging as a fundamentally novel type of treatment for various human diseases by addressing undruggable targets with existing medicines. Yet, free siRNA (small interfering RNA) injected into the blood exhibits a poor pharmacokinetic profile, as a consequence of the susceptibility of RNA molecules to serum nucleases, renal clearance, and non-specific biodistribution.1 Hence, the development of safe and effective delivery systems of siRNA is of crucial importance for a successful RNAi-based therapy.2,3In recent decade, great progress has been made in developing effective siRNA delivery systems. Among them, lipid-based nanoparticles are the most intensively investigated for systemic delivery of siRNA therapeutics. For improvement in the siRNA therapeutic efficacy, tremendous strategies have been explored to develop the siRNA nanoparticle formulations to protect the siRNA molecules from enzymatic degradation by endogenous nucleases after injection into a periphery vein, to avoid aggregation with both blood and extracellular elements and the subsequent uptake by phagocytes, to reduce immune response, and to enhance accumulation at target tissues. Until now, PEGylation is one of the most commonly used strategies in designing vector for preventing serum protein binding,4,5 aiding the formation of uniform nanoparticles,6 prolonging half-life in blood circulation,7 enhancing permeability and retention effect (EPR),7 and reducing mononuclear phagocyte.8,9 All these advantages may help overcome the barriers in developing lipid-based nanoparticles for in vitro and in vivo siRNA delivery.
Bicelles are an attractive category of versatile and robust lipid-detergent assemblies which have been described as bicellar nanodiscs.10 They are typically prepared from a mixture of a long chain phospholipid of dimyristoylphosphatidylcholine (DMPC) and a short chain phospholipid of 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC)11 or the bile-salt derivative of 3-(cholamidopropyl)dimethylammonio-2-hydroxy-1-propanesulfonate (CHAPSO).12 This bicellar nanodiscs technology represents a breakthrough in our capability to investigate the structure of various membrane proteins13 and function of membrane associated proteins.14 More importantly, the intrinsic versatility of this bicellar nanodiscs allows for loading a variety of therapeutic and diagnostic agents, such as doxorubicin,15 porphyrin,16 Gd-DTPA and Cy5,17 and siRNA.18 Such bicellar nanodiscs can serve as a powerful vehicle to address a variety of challenges faced by the drug and gene delivery since it combines the advantages of both lipid vesicles and the classical mixed micelles.19 Nevertheless, the bicellar nanodiscs may be destabilized upon subjecting to electrostatic, hydrophobic, and van der Waals interactions with plasma proteins. It was found that the bicellar nanodiscs could spontaneously transform into rod-like micelles or vesicles because of the elevation of temperature.20 The instability may limit the use of bicellar nanodiscs for drug and siRNA delivery. Robert et al. have assembled bicellar nanodiscs from apolipoprotein A-I, DMPC and 1,2-dimyristoyl-3-trimethylammoniumpropane (DMTAP). The bicellar nanodiscs exhibited 60% gene knock down efficiency,21 but self-association in the presence of siRNA have greatly increased the size of the complex which might not be suited for in vivo application. Therefore, there is pressing need to develop more stable bicellar nanodiscs for biomedical applications. In order to overcome general problems relating to current phospholipid bicellar nanodiscs, Kikuchi et al. have recently fabricated a new type of organic–inorganic hybrid bicellar nanodiscs from cerasome-forming lipid (CFL) and dihexanoyl phosphatidylcholine (DHPC) at the ratio of 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
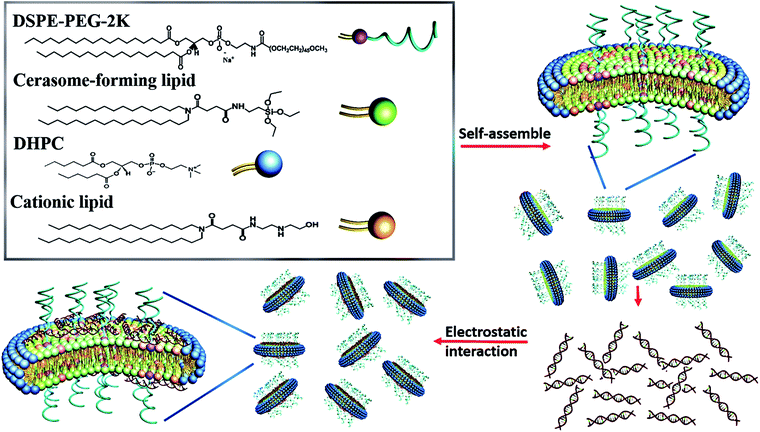
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.22 This hybrid nanodisc has attracted research interests because its nontoxic polyorganosiloxane surface may protect the inner lipid bilayer and impart remarkably higher stability than conventional phospholipid bicelles.23 In addition, the polyorganosiloxane surface is facile to conjugate with various ligands through silane-coupler chemistry for targeting delivery. The current study reports the development of the cationic PEGylated bicellar nanodiscs for the use as siRNA delivery vector by using conventional Bangham method in combination with sol–gel reaction and self-assembly process from cerasome-forming lipid of N-[N-(3-triethoxysilyl) propylsuccinamoyl]dihexadecylamine and a hydroxylated cationic lipid (HCL) of N1,N1-dihexadecyl-N4-(2-(2-hydroxyethylamino)ethyl)-succinamide, together with detergent DHPC and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-methoxy(polyethylene glycol)-2000 (DSPE-PEG2K) (Scheme 1). The PEGylation of the cationic bicellar nanodiscs by incorporating DSPE-PEG2K could protect the nanodiscs from agglomeration and macrophage capture, reduce protein absorption, and consequently prolong the blood circulating time. We investigated the physicochemical properties, biocompatibility, intracellular trafficking behavior and in vitro transfection efficiency of these hybrid bicellar nanodiscs. The siRNA delivery efficacy of the nanodiscs was assayed in vitro and in vivo as well. The unique characteristics of the cationic PEGylated hybrid bicellar nanodiscs address many of the deficiencies relating to current liposome technology.
2.22 This hybrid nanodisc has attracted research interests because its nontoxic polyorganosiloxane surface may protect the inner lipid bilayer and impart remarkably higher stability than conventional phospholipid bicelles.23 In addition, the polyorganosiloxane surface is facile to conjugate with various ligands through silane-coupler chemistry for targeting delivery. The current study reports the development of the cationic PEGylated bicellar nanodiscs for the use as siRNA delivery vector by using conventional Bangham method in combination with sol–gel reaction and self-assembly process from cerasome-forming lipid of N-[N-(3-triethoxysilyl) propylsuccinamoyl]dihexadecylamine and a hydroxylated cationic lipid (HCL) of N1,N1-dihexadecyl-N4-(2-(2-hydroxyethylamino)ethyl)-succinamide, together with detergent DHPC and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-methoxy(polyethylene glycol)-2000 (DSPE-PEG2K) (Scheme 1). The PEGylation of the cationic bicellar nanodiscs by incorporating DSPE-PEG2K could protect the nanodiscs from agglomeration and macrophage capture, reduce protein absorption, and consequently prolong the blood circulating time. We investigated the physicochemical properties, biocompatibility, intracellular trafficking behavior and in vitro transfection efficiency of these hybrid bicellar nanodiscs. The siRNA delivery efficacy of the nanodiscs was assayed in vitro and in vivo as well. The unique characteristics of the cationic PEGylated hybrid bicellar nanodiscs address many of the deficiencies relating to current liposome technology.
 | ||
| Scheme 1 Schematic illustration of the formation of the PEGylated bicellar nanodisc for siRNA delivery. | ||
Results and discussion
Preparation and characterization of bicellar nanodiscs
The PEGylated cationic hybrid bicellar nanodiscs (NDs) were fabricated from the mixture of CFL, HCL and DHPC at two designated molar ratios of 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7
7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 and 7
8 and 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7
7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 by introducing 0, 1, 3 and 5 mol% DSPE-PEG2K, respectively (Table 1).
4 by introducing 0, 1, 3 and 5 mol% DSPE-PEG2K, respectively (Table 1).
| Nanodiscs | Molar ratio CFL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CHL CHL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DHPC DHPC |
Mol% DSPE-PEG2K | Diameter (nm) by DLS | PDI | Zeta potential (mV) | |
|---|---|---|---|---|---|---|
| Before siRNA binding | After siRNA bindinga | |||||
a The results are obtained when the molar ratio of the cationic lipid nitrogen (N) to siRNA phosphate (P) is 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. 1. |
||||||
| ND1 | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 07 07![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 08 08 |
0 | 126.8 ± 29.29 | 0.161 ± 0.030 | 35.44 ± 0.88 | −9.72 ± 0.49 |
| ND2 | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 07 07![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 08 08 |
1 | 117.5 ± 21.50 | 0.128 ± 0.025 | 39.37 ± 0.48 | −9.74 ± 0.39 |
| ND3 | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 07 07![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 08 08 |
3 | 60.8 ± 3.53 | 0.184 ± 0.010 | 32.59 ± 0.44 | −8.53 ± 0.36 |
| ND4 | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 07 07![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 08 08 |
5 | 54.0 ± 2.59 | 0.181 ± 0.009 | 34.93 ± 0.33 | −4.81 ± 0.63 |
| ND5 | 07![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 07 07![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 04 04 |
0 | 63.9 ± 5.30 | 0.170 ± 0.011 | 32.29 ± 0.64 | −2.97 ± 0.32 |
| ND6 | 07![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 07 07![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 04 04 |
1 | 78.3 ± 3.84 | 0.251 ± 0.008 | 39.19 ± 0.82 | −5.68 ± 0.42 |
| ND7 | 07![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 07 07![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 04 04 |
3 | 53.5 ± 1.07 | 0.276 ± 0.012 | 42.84 ± 0.62 | −9.05 ± 0.68 |
| ND8 | 07![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 07 07![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 04 04 |
5 | 30.5 ± 3.81 | 0.245 ± 0.014 | 37.55 ± 0.44 | −7.31 ± 0.62 |
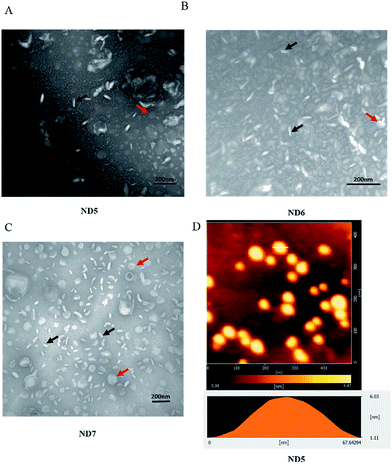
In order to obtain the discoidal structure, the molar ratio of the long chain lipids (CFL and HCL) to the short chain lipid of DHPC was kept at a constant of 3.5.24 The polyorganosiloxane coating on the NDs was formed by in situ sol–gel processes (Si–OCH2CH3 + H2O → Si–OH + CH3CH2OH followed by 2Si–OH → Si–O–Si + H2O). The morphology of the NDs was analyzed by transmission electron microscopy (TEM) and atomic forcing microscopy (AFM). As observed in the images (Fig. 1A–C), ND5, ND6 and ND7 appeared to be disc-like shape, and rod-like and disc-like objects were corresponding to the edge-on and face-on NDs, respectively. The AFM image showed that the size of ND5 ranged from 28.99 nm to 51.15 nm, with an average particle diameter of 37.54 ± 6.92 nm (based on 20 particles) (Fig. 1D), in agreement with the TEM measurements (Fig. 1A), and the thickness of the edge-on bicellar nanodiscs was evaluated to be 4.92 nm by AFM measurement further confirm the disc-like shape. The hydrodynamic size of all the NDs measured by dynamic light scattering (DLS) showed in (Table 1) were larger than the particle size determined by TEM and AFM (Fig. 1) owing to the fact that the DLS analyzes the mean diameter calculated from the diffusional properties of dynamic nonspherical NDs in the hydrated state, whereas TEM and AFM analyzes the NDs in the dried state.25,26 These hybrid NDs could keep their discoidal structure in a dry environment, indicating higher stability than conventional phospholipid bicelles which would rupture to form ambiguous aggregates while exposing to air.22
As we know, the PEG coating onto the gene nanovectors could prolong its circulation time but the gene transfection efficiency could be compromised. As observed in (Fig. 2A), NDs of ND1 to ND4 containing less HCL exhibited more obvious siRNA binding decrease along with the increase of PEG amount, 1 mol% PEG mixing had already increased the siRNA binding to N/P ratio of 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and further raise the PEG ratio decreased the binding efficiency dramatically. But the PEGylation had less effect on NDs of ND5 to ND8 which contain more HCL, this might contribute to the increased HCL which had offered more positive charge to weaken the shielding effect in a certain extent. But when PEG ratio increased to 5 mol%, the siRNA binding efficiency increase to N/P ratio of 8
1, and further raise the PEG ratio decreased the binding efficiency dramatically. But the PEGylation had less effect on NDs of ND5 to ND8 which contain more HCL, this might contribute to the increased HCL which had offered more positive charge to weaken the shielding effect in a certain extent. But when PEG ratio increased to 5 mol%, the siRNA binding efficiency increase to N/P ratio of 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 which was much higher than our previous work.27 Our explanation could be that for NDs, the PEG chains were all exposed on the surface of bilayer, but to the spherical liposome structure, PEG chains were exposed on both the outer and the inner surface. The structure differences subsequently resulted in distinct PEGylation induced steric shielding effect, accordingly, with equivalent PEG containing lipid mix ratio, NDs presented more obvious PEG-associate steric shielding effect.
1 which was much higher than our previous work.27 Our explanation could be that for NDs, the PEG chains were all exposed on the surface of bilayer, but to the spherical liposome structure, PEG chains were exposed on both the outer and the inner surface. The structure differences subsequently resulted in distinct PEGylation induced steric shielding effect, accordingly, with equivalent PEG containing lipid mix ratio, NDs presented more obvious PEG-associate steric shielding effect.
The phosphate groups of siRNA would neutralize the NH3+ groups of cationic lipid therefore lower the positive charges of the cationic hybrid NDs, and the zeta potential of NDs/siRNA complexes depend on the ratio of the positive charges of cationic lipid to the number of phosphate groups of siRNA. Taken the gel electrophoresis data as a reference, we characterized the NDs/siRNA complexes at N/P ratio of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. As shown in (Table 1), zeta potential of the NDs/siRNA complexes were all negative, which indicates successful association of siRNA with the NDs. The slight negative charge on the NDs/siRNA complexes may be beneficial for in vivo delivery because though the positive surface charge facilitates carriers to bind to the negatively-charged cell surface, excessive positive charge can lead to nonspecific binding and significant toxicity.28 And in complicated physiological environment, negatively-charged serum proteins in the bloodstream is more likely to bind to positively-charged complexes, rendering it ineffective.29 Therefore, NDs/siRNA complexes with N/P ratio of 4
1. As shown in (Table 1), zeta potential of the NDs/siRNA complexes were all negative, which indicates successful association of siRNA with the NDs. The slight negative charge on the NDs/siRNA complexes may be beneficial for in vivo delivery because though the positive surface charge facilitates carriers to bind to the negatively-charged cell surface, excessive positive charge can lead to nonspecific binding and significant toxicity.28 And in complicated physiological environment, negatively-charged serum proteins in the bloodstream is more likely to bind to positively-charged complexes, rendering it ineffective.29 Therefore, NDs/siRNA complexes with N/P ratio of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 were selected for further investigation in vitro and in vivo.
1 were selected for further investigation in vitro and in vivo.
Cytotoxicity and silencing efficacy of NDs/siRNA complexes in vitro
The cytotoxicity of NDs and NDs/siRNA complexes was evaluated in a hepatocellular carcinoma-derived luciferase-expressing (HepG2-Luc) cell line, using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay. As shown in (Fig. S1†), the mixing of CFL effectively decreased the cytotoxicity of naked carriers. And as observed in (Fig. 2B), the viability of NDs/siRNA complexes are all over 80% which indicates negligible in vitro cytotoxicity.30 The gene silencing efficiency of the NDs was assessed with targeting luciferase siRNA on HepG2-Luc cell line. A commercially available transfection reagent, Lipofectamine 2000 was chosen as positive control. As shown in (Fig. 2C), all NDs/siRNA complexes showed certain firefly luciferase-expression inhibition and the ND5 even realized equivalent gene silencing efficiency with Lipofectamine 2000. Notably, comparing to ND5, the silencing efficiency of ND6 with 1 mol% PEG was slightly lower. But the silencing efficiency dropped obviously when PEG ratio was up to 3 mol%. Combining with the gel retardation result we can find that excess PEG will cause reduced siRNA binding efficiency which lead to the decreased transfection efficiency.Quantitative real-time polymerase chain reaction (qRT-PCR) was applied to further confirm the silencing efficiency. Herein, Plk1 gene was chosen as target gene since it shows elevated activity in various cancers and is a key regulator of mitotic progression in mammalian cells.31,32 Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was employed as internal control as it is highly conserved. As shown in (Fig. 2D), all the ND/siPlk1 complexes down-regulated Plk1 gene expression in cells. Still, the qRT-PCR result of silencing efficiency of ND5 was equivalent to Lipofectamine 2000 group which was in accordance with the luciferase assay.
Cellular internalization of NDs/siRNA complexes
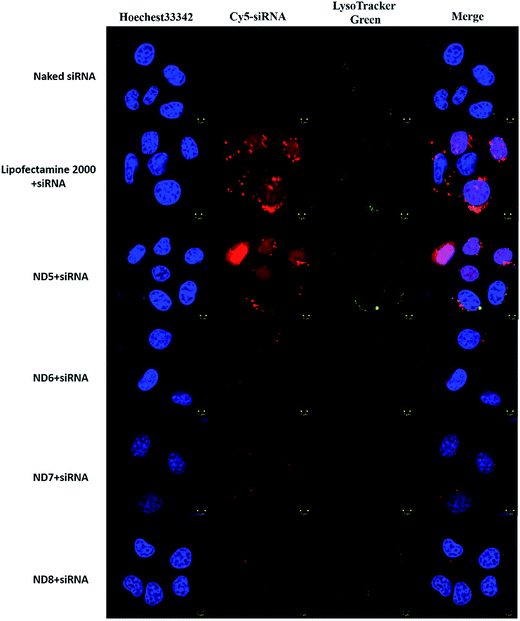
The subcellular distribution of NDs/Cy5-labelled siRNA was observed with confocal laser scanning microscope (CLSM). Hoechst 33342 was used to stain nuclei and LysoTracker®Green was used to visualize the endosomes. As shown in (Fig. 3A and S2†), in the naked siRNA treated group, no obvious Cy5 fluorescence signal in the cytoplasm were observed after 4 h incubation due to the poor cellular uptake.33 Comparing to the naked group, the NDs/siRNA complexes effectively delivered siRNA into cells, as strong Cy5 signal was observed in the cytosol, especially for ND5/siRNA group, which showed similar pattern with lipofectamine 2000. It is obvious that, with the increase of PEG mix ratio, ND6, ND7 and ND8/siRNA complexes showed gradually decreased red signal intensity, this phenomenon was attributed to the PEGylation caused steric shielding effect which lead to the decreased siRNA binding efficiency and cellular uptake. In previous work,27 the cationic cerasomes with 5 mol% PEG decreased the cellular uptake but not as obviously as the NDs. Comparing to the spherical shape cationic cerasomes which contain HCL, CFL, and 5 mol% DSPE-PEG2K, the only distinction of the components of NDs is the DHPC which causes the shape difference. But the same PEG ratio had caused totally different results, indicating that the exposed PEG amount caused by shape difference of the NDs and cationic cerasomes have great impact on the cellular uptake. And again, this finding revealed that excess PEG will lead to reduced cellular uptake.Subsequently, the internalizations of all these NDs/Cy5-labelled siRNA complexes were measured by fluorescence-activated cell sorting (FACS) (Fig. 4). In accordance with the results of confocal observation (Fig. 3), ND5 showed similar cellular uptake as Lipofectamine 2000, while the increasing PEG in ND6-8 leaded to lower cellular uptake, which is also in accordance with silencing efficiency (Fig. 2C and D).
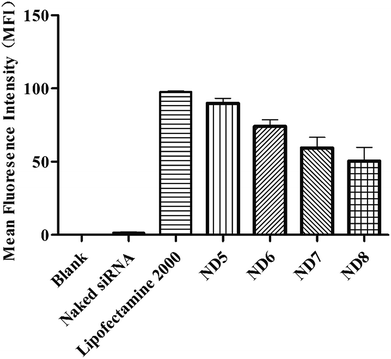 | ||
| Fig. 4 The internalizations of NDs/Cy5–siRNA complexes were measured by fluorescence-activated cell sorting (FACS) and their mean fluorescence intensities were showed. | ||
In vivo distribution of NDs/siRNA
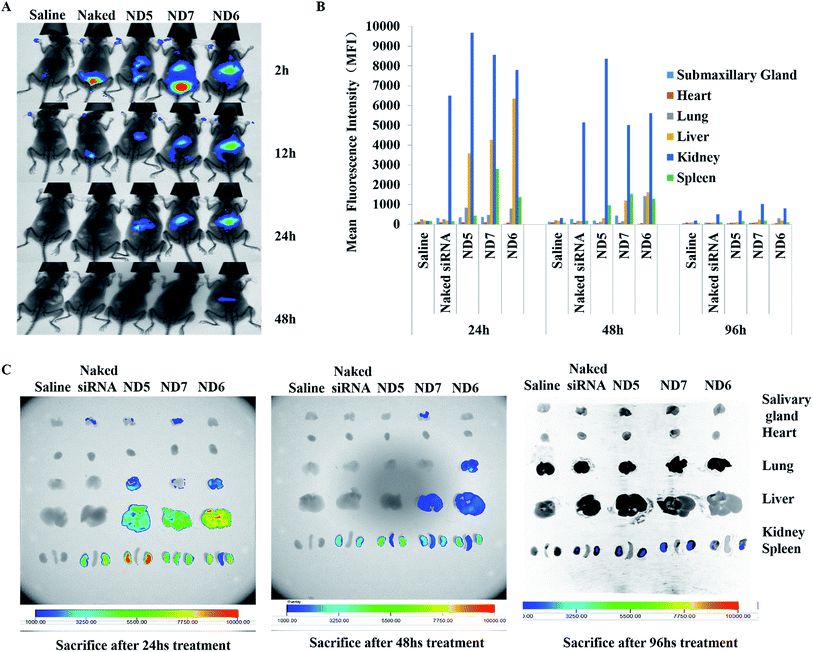
To demonstrate the in vivo distribution of the nanodiscs, we administrated ND5/siRNA, ND6/siRNA and ND7/siRNA complexes (Cy5–siRNA) by tail vein injection to C57BL/6 mice and monitored the distributions by whole-body fluorescence imaging (Fig. 5A). Similar to the reported results,34–38 naked siRNA experienced rapid renal clearance upon systemic administration. Isolated organ fluorescence intensity analysis (Fig. 5B and C) further demonstrated that naked siRNA exhibited much weaker signal intensity in liver, lung, spleen but the NDs/siRNA groups exhibited strong MFIs. However, all mice administered siRNA had comparable fluorescence intensity in the submandibular gland and kidney. This suggested that the NDs have potential application in delivering siRNA to liver, lung or spleen to cure diseases, especially cancer. As observed in (Fig. 5), the PEGylated ND6 and ND7 showed slower clearance efficiency and gradual fluorescence fading than the non-PEGylated ND5 after administration proved the prolonged blood circulation effect of PEG.It is worth noting that ND7 with 3 mol% PEG showed less accumulation in liver than ND6 with 1 mol% PEG. This result suggested that although the PEGylation can prolong the circulation time in vivo, the decreased siRNA binding and cellular uptake caused by PEG shielding effect will lead to poor organ accumulation. With fully expose the PEG chain outside the surface of the nanoparticles, 1 mol% PEG was enough to prolong the blood circulation effectively without compromising the transfection efficiency and organ accumulation.
Conclusions
We prepared hybrid cationic bicellar nanodiscs (NDs) composed of HCL, CFL, DHPC and DSPE-PEG2K with good stability and biocompatibility. The modest siRNA delivery efficiency in vitro and in vivo makes the hybrid bicellar nanodiscs a promising platform for developing siRNA delivery vehicles. Additionally, we have revealed that cellular uptake affected by the PEG shielding was closely related with the PEG ratio of NDs. And the in vivo distribution and isolated organ analysis suggested that although PEGylation enhanced circulation retention but excess PEG had led to poor organ accumulation. Hence, if PEGylation was used for prolong the circulation time, multi mix ratio should be tested to select the optimized ratio which would efficiently improve the in vivo retention time without sacrificing the transfection efficiency. And the shape of the nanoparticles which will decide the expose amount of PEG should be carefully considered.Experimental
Materials
1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine-N-methoxy (polyethylene glycol)-2000 (DSPE-PEG2K) was purchased from Shanghai Advanced Vehicle Technology Pharmaceutical Ltd. Other organic reagents or solvents were of analytical grade. All the chemicals were used directly without further purification. The cerasome-forming lipid (CFL) was synthesized following the previous reported.39 The synthesis procedure and characterization of hydroxylated cationic lipid of N1,N1-dihexadecyl-N4-(2-(2-hydroxyethylamino)ethyl) succinamide (HCL) was synthesized according to the literature.27 The siLuc (targeting firefly luciferase gene) and siPlk1 (targeting polo-like kinase 1 gene) were supplied by Suzhou Ribo Life Science Co. (Jiangsu, China). The sequences were as follows: siLuc-sense: 5′-CCCUAUUCUCCUUCUUCGCdTdT-3′, siLuc-antisense: 5′-GCGAAGAAGGAGAAUAGGGdTdT-3′; Cy5-NC-sense: 5′-Cy5-CCUUGAGGCAUACUUCAAAdTdT-3′, Cy5-NC-antisense: 5′-UUUGAAGUAUGCCUCAAGGdTdT-3′; siPlk1-sense: 5′-UGAAGAAGAUCACCCUCCUUAdTdT-3′, siPlk1-antisense: 5′-CAGAAGUCAUCUUGAUAGAdTdT-3′; Hochest 33342 was from Dojindo Molecular Technologies, Inc. Lipofectamine 2000 was purchased from Invitrogen. Dulbecco' modified Eagle medium (DMEM), MTT, Opti-MEM, penicillin/streptomycin and trypsin were from Hyclone.Preparation of PEGylated cationic hybrid NDs
The NDs were prepared according to the methods described by ref. 40. Briefly, HCL, CFL and DHPC were mixed in deionized water to achieve a final lipid concentration of 3 mg mL−1, followed by a series of cycles of freezing, thawing, and gentle vortexing until the turbid mixture became transparent solution, indicating NDs had formed. The suspension was stored at 4 °C for up to 24 h before use to enable the formation of polyorganosiloxane surface. For comparative investigation, NDs with 5 mol%, 3 mol%, 1 mol% DSPE-PEG2K were prepared using the same procedure.Characterization of NDs/siRNA complexes
Preparation of NDs/siRNA complexes and gel retardation assay
For siRNA binding, NDs diluted at different concentrations with diethyl pyrocarbonate (DEPC) water were mixed with desired amount of siRNA solution (20 μM) by gentle pipetting and incubated at room temperature for 20 min before characterization. Electrophoresis was performed on 2% agarose gel containing 1 μL Gelsafe per 20 mL 2% agarose gel at the voltage of 90 V for 20 min in Tris-acetate-EDTA (TAE) running buffer solution. The nucleic acid bands were visualized by UV imaging equipment (ABI, GIS-2500).Cytotoxicity measurements
The cytotoxicity of the NDs and NDs/siRNA complexes were assessed by a MTT viability assay against HepG2-Luc cells. Cells were seeded in 96-well plates at 1.0 × 104 cells per well and cultured under the environment mentioned above overnight, then cells were separately incubated with different concentrations of NDs or NDs/siRNA complexes and further cultured for 24 h. Cells used as control were treated with an equivalent volume of PBS treatment. Then, MTT stock solution was added into each well to achieve a final concentration of 1 g L−1 and the plate was incubated for an additional 4 h, after which all medium was removed and 150 μL of the DMSO (cell degrade) was added into each well and further incubated in dark for 15 min. Finally, the absorbance was read at 570 nm with a reference wavelength of 630 nm using a Microplate reader (Synergy HT, BioTek, USA). Cell viability was normalized to cells maintained in complete DMEM with PBS treatment.In vitro gene silencing
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, and the supernatant was collected for luminescence measurements. The relative light units (RLUs) were measured with a fluorometer (Synergy HT, BioTek, USA).
000 rpm, and the supernatant was collected for luminescence measurements. The relative light units (RLUs) were measured with a fluorometer (Synergy HT, BioTek, USA).qRT-PCR analysis
The cells were transfected using the method above and total cellular RNA was isolated using an RNeasy mini kit (Qiagen, Valencia, CA) according to the manufacturer's protocol to perform the qRT-PCR analysis. cDNA was synthesized using the PrimeScript First Strand cDNA Synthesis Kit (Promega, A3500, USA) with two microgram of total RNA. Then, the reaction mixture was subjected to qRT-PCR which was performed using the Applied Biosystems StepOne Real-Time PCR Systems. The housekeeping gene GAPDH was used as an internal standard. The primers used in the qRT-PCR for Plk1 and GAPDH were as follow: Plk1-forward: 5′-GCCCCTCACAGTCCTCAATA-3′; Plk1-reversed: 5′-TACCCAAGGCCGTACTTGTC-3′; GAPDH-forward: 5′-AGAAGGCTGGGGCTCATTTG-3′; GAPDH-reversed: 5′-AGGGGCCATCCACAGTCTTC-3′.Cellular internalization of NDs/Cy5-siRNA
HepG2-Luc cells were seeded in 35 mm dish at 2 × 105 cells per well and cultured for 24 h at 37 °C. After 24 h, cells were transfected with complexes containing 50 nM (final concentration) Cy5-labelled siRNA in Opti-MEM for 4 h at 37 °C. Then the treated cells were washed three times with 1 mL PBS to remove residual free complexes. Meanwhile, LysoTracker®Green DND-26 (Invitrogen, Carlsbad, CA) was used to indicate the endosome/lysosome organelles, and Hoechst 33342 was used to stain cell nuclei. Distributions of the Cy5-labelled siRNA were examined and recorded in living cells by using Zeiss confocal microscopy (LSM 700, Carl Zeiss, Germany). Imaging processing programs were coded in Interactive Data Language.Cellular uptake measured by fluorescence-activated cell sorting (FACS)
In order to estimate cellular uptake efficiency of complexes, HepG2-Luc cells were seeded in 6-well plates at 2 × 105 cells per well and cultured for 24 h at 37 °C. After 24 h, cells were transfected with complexes containing 50 nM (final concentration) Cy5-labelled siRNA in Opti-MEM for 4 h at 37 °C. Then the treated cells were washed three times with 1 mL PBS to remove residual free complexes, and suspended in PBS, subsequently introduced into a FACS Calibur flow cytometer (Becton Dickinson, San Jose, CA, USA). Median fluorescence intensity was given.In vivo distribution of NDs/Cy5-siRNA
Female C57BL/6 mice, 8–10 weeks old, weighing 19–20 g were purchased from Vital River Co. (Beijing, China). Animals were maintained in Peking University Laboratory Animal Center (an Association for Assessment and Accreditation of Laboratory Animal Care-accredited experimental animal facility). All procedures involving experimental animals were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of Peking University and Guide for the Care and Use of Laboratory Animals approved by National Research Council in 1996.For in vivo distribution, a given formulation of Cy5-labelled siRNA was administered to each mouse via subcutaneous injection at 2 mg kg−1. The Cy5 fluorescence signal from the whole body was detected using a Kodak in vivo imaging system (Kodak In Vivo Imaging System FX Pro, Carestream Health, USA) at given time points. At the endpoint, mice were sacrificed by cervical dislocation, and certain organs and tissue were isolated and examined using the Kodak in vivo imaging.
Statistical analysis
Student's t-test was used to assess the statistical significance of the experimental results. P values of 0.05 or less than 0.05 were considered statistically significant, while values of 0.01 or less were considered extreme significant.Acknowledgements
This work was financially supported by National Key Research and Development Program of China (No. 2016YFA0201400), State Key Program of National Natural Science of China (Grant No. 81230036), National Natural Science Foundation for Distinguished Young Scholars (No. 81225011), National Natural Science Foundation of China (No. 81473128, No. 81371580) and the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (No. 81421004). National Basic Research Program of the Chinese Ministry of Science and Technology (973 Grant No. 2013CB531202).Notes and references
- K. A. Whitehead, R. Langer and D. G. Anderson, Nat. Rev. Drug Discovery, 2010, 9, 412 CrossRef.
- A. Wittrup and J. Lieberman, Nat. Rev. Genet., 2015, 16, 543–552 CrossRef CAS PubMed.
- H. Yin, R. L. Kanasty, A. A. Eltoukhy, A. J. Vegas, J. R. Dorkin and D. G. Anderson, Nat. Rev. Genet., 2014, 15, 541–555 CrossRef CAS PubMed.
- T. Shimizu, Y. Mima, Y. Hashimoto, M. Ukawa, H. Ando, H. Kiwada and T. Ishida, Immunobiology, 2015, 220, 1151–1160 CrossRef CAS PubMed.
- S. H. Hsu, B. Yu, X. Wang, Y. Lu, C. R. Schmidt, R. J. Lee, L. J. Lee, S. T. Jacob and K. Ghoshal, Nanomedicine: Nanotechnology, Biology and Medicine, 2013, 9, 1169–1180 CrossRef CAS PubMed.
- Q. Xu, L. M. Ensign, N. J. Boylan, A. Schon, X. Gong, J. C. Yang, N. W. Lamb, S. Cai, T. Yu, E. Freire and J. Hanes, ACS Nano, 2015, 9, 9217–9227 CrossRef CAS PubMed.
- D. Vllasaliu, R. Fowler and S. Stolnik, Expert Opin. Drug Delivery, 2014, 11, 139–154 CrossRef CAS PubMed.
- Y. Li, R. Lin, L. Wang, J. Huang, H. Wu, G. Cheng, Z. Zhou, T. MacDonald, L. Yang and H. Mao, J. Mater. Chem. B, 2015, 3, 3591–3603 RSC.
- M. L. Immordino, F. Dosio and L. Cattel, Int. J. Nanomed., 2006, 1, 297–315 CrossRef CAS PubMed.
- C. R. Sanders and R. S. Prosser, Structure, 1998, 6, 1227–1234 CrossRef CAS PubMed.
- N. Matsumori, A. Morooka and M. Murata, J. Am. Chem. Soc., 2007, 129, 14989–14995 CrossRef CAS PubMed.
- C. R. Sanders 2nd and J. H. Prestegard, Biophys. J., 1990, 58, 447–460 CrossRef CAS.
- Y. Gao, E. Cao, D. Julius and Y. Cheng, Nature, 2016, 534, 347–351 CrossRef CAS PubMed.
- T. Yamaguchi, T. Uno, Y. Uekusa, M. Yagi-Utsumi and K. Kato, Chem. Commun., 2013, 49, 1235–1237 RSC.
- W. Zhang, J. Sun, Y. Liu, M. Tao, X. Ai, X. Su, C. Cai, Y. Tang, Z. Feng, X. Yan, G. Chen and Z. He, Mol. Pharm., 2014, 11, 3279–3290 CrossRef CAS PubMed.
- K. K. Ng, J. F. Lovell, A. Vedadi, T. Hajian and G. Zheng, ACS Nano, 2013, 7, 3484–3490 CrossRef CAS PubMed.
- R. Duivenvoorden, J. Tang, D. P. Cormode, A. J. Mieszawska, D. Izquierdo-Garcia, C. Ozcan, M. J. Otten, N. Zaidi, M. E. Lobatto, S. M. van Rijs, B. Priem, E. L. Kuan, C. Martel, B. Hewing, H. Sager, M. Nahrendorf, G. J. Randolph, E. S. Stroes, V. Fuster, E. A. Fisher, Z. A. Fayad and W. J. Mulder, Nat. Commun., 2014, 5, 3065 Search PubMed.
- M. Ghosh, G. Ren, J. B. Simonsen and R. O. Ryan, Biochem. Cell Biol., 2014, 92, 200–205 CrossRef CAS PubMed.
- I. G. Denisov, Y. V. Grinkova, A. A. Lazarides and S. G. Sligar, J. Am. Chem. Soc., 2004, 126, 3477–3487 CrossRef CAS PubMed.
- L. van Dam, G. Karlsson and K. Edwards, Biochim. Biophys. Acta, 2004, 1664, 241–256 CrossRef CAS PubMed.
- M. Ghosh, G. Ren, J. B. Simonsen and R. O. Ryan, Biochem. Cell Biol., 2014, 92, 200–205 CrossRef CAS PubMed.
- K. Yasuhara, S. Miki, H. Nakazono, A. Ohta and J. Kikuchi, Chem. Commun., 2011, 47, 4691–4693 RSC.
- L. Lin, X. Y. Wang, Y. Y. Guo, K. Ren, X. D. Li, L. J. Jing, X. Yue, Q. Zhang and Z. F. Dai, RSC Adv., 2016, 79811–79821 RSC.
- M. P. Nieh, V. A. Raghunathan, C. J. Glinka, T. A. Harroun, G. Pabst and J. Katsaras, Langmuir, 2004, 20, 7893–7897 CrossRef CAS PubMed.
- K. G. Kulikov and T. V. Koshlan, Tech. Phys., 2015, 60, 1758–1764 CrossRef CAS.
- M. Y. Lee, S. J. Park, K. Park, K. S. Kim, H. Lee and S. K. Hahn, ACS Nano, 2011, 5, 6138–6147 CrossRef CAS PubMed.
- Y. Y. Li, S. Q. Zheng, X. L. Liang, Y. S. Jin, Y. D. Wu, H. C. Bai, R. F. Liu, Z. F. Dai, Z. C. Liang and T. J. Shi, Bioconjugate Chem., 2014, 25, 2055–2066 CrossRef CAS PubMed.
- M. Ogris, S. Brunner, S. Schuller, R. Kircheis and E. Wagner, Gene Ther., 1999, 6, 595–605 CrossRef CAS PubMed.
- K. A. Whitehead, R. Langer and D. G. Anderson, Nat. Rev. Drug Discovery, 2009, 8, 129–138 CrossRef CAS PubMed.
- H. Katas and H. O. Alpar, J. Controlled Release, 2006, 115, 216–225 CrossRef CAS PubMed.
- K. Strebhardt and A. Ullrich, Nat. Rev. Cancer, 2006, 6, 321–330 CrossRef CAS PubMed.
- R. E. Gutteridge, M. A. Ndiaye, X. Liu and N. Ahmad, Mol. Cancer Ther., 2016, 15, 1427–1435 CrossRef CAS PubMed.
- Y. Zhang, A. Satterlee and L. Huang, Mol. Ther., 2012, 20, 1298–1304 CrossRef CAS PubMed.
- F. Iversen, C. Yang, F. Dagnaes-Hansen, D. H. Schaffert, J. Kjems and S. Gao, Theranostics, 2013, 3, 201–209 CrossRef CAS PubMed.
- S. Gao, F. Dagnaes-Hansen, E. J. Nielsen, J. Wengel, F. Besenbacher, K. A. Howard and J. Kjems, Mol. Ther., 2009, 17, 1225–1233 CrossRef CAS PubMed.
- J. Soutschek, A. Akinc, B. Bramlage, K. Charisse, R. Constien, M. Donoghue, S. Elbashir, A. Geick, P. Hadwiger, J. Harborth, M. John, V. Kesavan, G. Lavine, R. K. Pandey, T. Racie, K. G. Rajeev, I. Rohl, I. Toudjarska, G. Wang, S. Wuschko, D. Bumcrot, V. Koteliansky, S. Limmer, M. Manoharan and H. P. Vornlocher, Nature, 2004, 432, 173–178 CrossRef CAS PubMed.
- A. Santel, M. Aleku, O. Keil, J. Endruschat, V. Esche, G. Fisch, S. Dames, K. Loffler, M. Fechtner, W. Arnold, K. Giese, A. Klippel and J. Kaufmann, Gene Ther., 2006, 13, 1222–1234 CrossRef CAS PubMed.
- D. Bumcrot, M. Manoharan, V. Koteliansky and D. W. Sah, Nat. Chem. Biol., 2006, 2, 711–719 CrossRef CAS PubMed.
- K. Katagiri, M. Hashizume, K. Ariga, T. Terashima and J. Kikuchi, Chemistry, 2007, 13, 5272–5281 CrossRef CAS PubMed.
- R. Soong and P. M. Macdonald, Langmuir, 2009, 25, 380–390 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Cytotoxicity of naked NDs, intracellular distributions and cellular uptake of ND1–4/siRNA complexes. See DOI: 10.1039/c6ra24268e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2016 |