Effects of cytotoxicity of erythromycin on PAH-degrading strains and degrading efficiency
Abstract
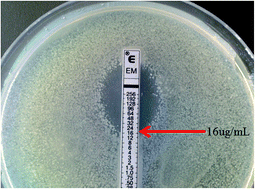
The toxicity and degradation effects of erythromycin on a phenanthrene degrading strain of Sphingomonas sp. GY2B and a pyrene degrading strain of Mycobacterium gilvum CP13 were investigated. The minimum inhibitory concentration (MIC) of erythromycin to GY2B was 0.25 mg L−1, but the MIC of erythromycin to CP13 was 16 mg L−1. Total superoxide dismutase (SOD) activity and lactate dehydrogenase (LDH) activity were significantly reduced when the erythromycin concentration exceeded the MIC. Erythromycin also reduced the growth of the strains and the biodegradation of polycyclic aromatic hydrocarbons (PAHs). Addition of erythromycin decreased GY2B phenanthrene removal efficiency by 0.81–86.03% compared to the control. Erythromycin also reduced CP13 pyrene degradation. The lowest pyrene removal efficiency was 10.78% when the erythromycin concentration was 256 mg L−1. The toxicity of erythromycin inhibited the growth of both GY2B and CP13.

 Please wait while we load your content...
Please wait while we load your content...