Investigation of twisted intercalating nucleic acid (TINA)-modified antisense oligonucleotides for splice modulation by induced exon-skipping in vitro†
Bao T. Leab,
Vyacheslav V. Filichevc and
Rakesh N. Veedu*ab
aCentre for Comparative Genomics, Murdoch University, Perth 6150, Australia. E-mail: r.veedu@murdoch.edu.au
bWestern Australian Neuroscience Research Institute, Perth 6150, Australia
cInstitute of Fundamental Sciences, Massey University, Palmerston North, 4442, New Zealand
First published on 27th September 2016
Abstract
Twisted intercalating nucleic acids (TINA)-modified oligonucleotides containing phenylethynylpyren-1-yl derivatives were reported to improve the thermal stability of the nucleic acid duplexes, triplexes and G-quadruplexes. In the present study, we have investigated the potential of TINA-modified antisense oligonucleotides (AOs) in splice modulation by induced exon-skipping in vitro. We used both para-TINA (p-TINA) and ortho-TINA (o-TINA)-modified 2′-O-methyl (2′-OMe) AOs on a phosphorothioate backbone (PS) designed to skip exon-23 in dystrophin pre-mRNA transcript in mdx mice myotubes. A fully-modified 2′-OMePS AO control was used in parallel. Our results showed that both p-TINA and o-TINA-modified AOs were able to induce exon-23 skipping. These results provide new insights on expanding the applicability of TINA-modified oligonucleotides.
Introduction
Antisense oligonucleotides (AOs) are prominent class of therapeutic nucleic acids that can regulate the expression of RNA and attracted considerable attention for drug development. AOs composed of natural nucleic acid monomers (DNA or RNA) possess relatively low binding affinity to complementary RNA or DNA sequences, and show very poor resistance to enzymatic degradation.1 To overcome this, chemically-modified nucleotide analogues are generally introduced to modify the nucleobase, sugar moiety or the internucleotide linkages.2,3 So far, two AOs have been approved by the United States Food and Drug Administration for the treatment of cytomegaloviral retinitis (Vitravene) and familial hypercholesterolemia (Kynamro).3 Twisted intercalating nucleic acid monomer possessing 1-ethynylpyrene moiety in the para position of the benzene ring (p-TINA, Fig. 1) was initially designed to increase thermal stability of Hoogsteen-type parallel DNA duplexes and triple helixes.4,5 Later, by changing the position of 1-ethynylpyrene moiety from para to ortho position of the benzene ring (o-TINA, Fig. 1), the thermal stability of Watson–Crick type antiparallel duplex helices was significantly improved.6 o-TINA molecule is now commercially available and generally used in constructing primer sequences for PCR amplification. AO-induced exon-skipping therapy in Duchene muscular dystrophy (DMD) has been studied extensively and advanced as the most promising therapy for DMD, a fatal muscle wasting disorder mainly affecting young boys, that leads to death in early adulthood.7–9 In this study, we have evaluated the potential of both p-TINA and o-TINA molecules modified antisense oligonucleotides to induce exon-skipping in DMD10 as model system.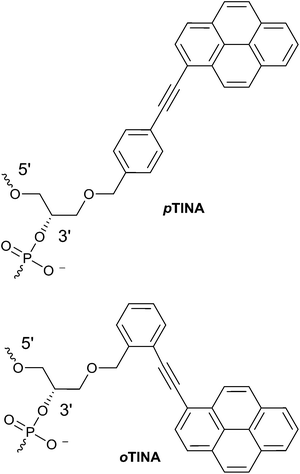 | ||
| Fig. 1 Structural representations of para (p)-TINA and ortho (o)-TINA monomers on a phosphodiester backbone. | ||
Materials and methods
Design and synthesis of 2′-OMePS antisense oligonucleotides
The 1p-TINA/2′-OMePS and 2′-OMePS AO (Table 1) were prepared in-house via standard phosphoramidite chemistry in 1 μmol scale on an expedite 8909 synthesiser as reported previously.11 The 1o-TINA/2′-OMePS and 2o-TINA/2′-OMePS AOs (Table 1) were purchased from Trilink Biotech.| Chemistries and AOs names | Sequence, 5′ → 3′ direction |
|---|---|
| a 2′-OMe nucleotides are represented in italic letter; ortho-TINA monomers are represented in bold underlined O. para-TINA monomer is represented in bold underlined P. All 2′-OMe and TINA-modified oligonucleotides possess phosphorothioate backbone (PS). | |
| DNA | d(GGCCAAACCTCGGCTTACCT) |
| RNA | r(GGCCAAACCUCGGCUUACCU) |
| 2′-OMePS | GGCCAAACCUCGGCUUACCU |
| 1p-TINA/2′-OMePS | ![[P with combining low line]](https://www.rsc.org/images/entities/b_char_0050_0332.gif) GGCCAAACCUCGGCUUACCU GGCCAAACCUCGGCUUACCU |
| 1o-TINA/2′-OMePS | ![[O with combining low line]](https://www.rsc.org/images/entities/b_char_004f_0332.gif) GGCCAAACCUCGGCUUACCU GGCCAAACCUCGGCUUACCU |
| 2o-TINA/2′-OMePS | ![[O with combining low line]](https://www.rsc.org/images/entities/b_char_004f_0332.gif) GGCCAAACCUCGGCUUACC GGCCAAACCUCGGCUUACC![[O with combining low line]](https://www.rsc.org/images/entities/b_char_004f_0332.gif) U U |
Melting temperature study of the antisense oligonucleotides
UV-Vis spectroscopy was performed using a Cary 100Bio UV-Vis spectrometer and quartz cuvettes with 1 cm path length and a 2 × 6 multicell block with a Peltier temperature controller. Extinction coefficient (ε) of TINA is 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 [L (mol × cm)−1]. DNA melting profiles were recorded by monitoring absorbance at 260 nm using 1 °C min−1 temperature ramp. Duplexes were prepared at 2.0 μM strand concentrations in Na+ phosphate buffer (5 mM) supplemented with 70 mM NaCl and 0.05 mM Na2-EDTA. The melting temperatures (Tm [°C]) were determined as the maxima of the first derivative plots of the melting curves.
000 [L (mol × cm)−1]. DNA melting profiles were recorded by monitoring absorbance at 260 nm using 1 °C min−1 temperature ramp. Duplexes were prepared at 2.0 μM strand concentrations in Na+ phosphate buffer (5 mM) supplemented with 70 mM NaCl and 0.05 mM Na2-EDTA. The melting temperatures (Tm [°C]) were determined as the maxima of the first derivative plots of the melting curves.
In vitro stability study of antisense oligonucleotides
Nuclease stability of the oligonucleotides was assayed at 5 μM oligonucleotides concentration using 0.24 units/reaction of snake venom phosphodiesterase (Sigma) in a buffer of 10 mM Tris–HCl, pH 8.9, 100 mM NaCl, and 15 mM MgCl2 in a final volume of 60 μl. Briefly, 8 μl reaction aliquots were removed at specific time points and an equal volume of 80% formamide containing bromophenol blue and xylene cyanol gel tracking dyes was added followed by heating for 5 min at 95 °C. Products were then analysed by 20% denaturing polyacrylamide gel electrophoresis. Quantitation was performed on a Biorad Chemidoc XRS imaging system.Cell culture and transfection
H2K mdx mouse myoblasts were cultured in Dulbecco's Modified Eagle Medium (DMEM) containing 20% fetal bovine serum (FBS), 10% horse serum (HS) supplemented with 0.5% chicken embryo extract until reached 60–90% confluency. Prior to cell seeding, the 24-well plate was pre-treated with 50 μg ml−1 poly-D-lysine (Sigma) and 100 μg ml−1 Matrigel (Corning). Then, the myoblasts were seeded at 2 × 104 cells per well in DMEM 5% HS and incubated at 37 °C, 5% CO2 for 48 hours for differentiation into myotubes.12 For transfection, antisense oligonucleotides were complexed with Lipofectin (Life Technologies) at the ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Lipofectin
1 (Lipofectin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AO) with final concentrations at 100 nM and 200 nM in a volume of 500 μl per well as per the manufacturer's instructions without removing the medium after 3 hours.13 [H2K cells were gifted by Prof. Steve Wilton and Prof. Susan Fletcher Lab, Murdoch University; and the experiments were conducted with the approval according to the Institutional guidelines and regulations.]
AO) with final concentrations at 100 nM and 200 nM in a volume of 500 μl per well as per the manufacturer's instructions without removing the medium after 3 hours.13 [H2K cells were gifted by Prof. Steve Wilton and Prof. Susan Fletcher Lab, Murdoch University; and the experiments were conducted with the approval according to the Institutional guidelines and regulations.]
RNA extraction and reverse transcription-polymerase chain reaction (RT-PCR)
Twenty-four hours after transfection, the cells were collected for RNA extraction. Briefly, RNA was extracted using Direct-zol™ RNA MiniPrep Plus with TRI Reagent® (Zymo Research) as per the manufacturer's instructions. The dystrophin transcripts were then amplified by nested RT-PCR across exons 20–26 as described previously.13 PCR products were separated on 2% agarose gels in Tris-acetate–EDTA buffer and the images were captured on a Fusion Fx gel documentation system (Vilber Lourmat, Marne-la-Vallee, France). Densitometry was performed by Image J software.Cell toxicity assay
Cells were seeded and transfected with AOs as described previously.12,13 After 24 h, cell viability was measured by WST-1 assay kit (Sigma). Briefly, WST-1 solution was added at ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (v/v) per well and incubated for 2 h at 37 °C, 5% CO2. The absorbance was measured with a microplate reader (FLUOstar Omega, BMG Labtech, Germany) at 450 nm.
10 (v/v) per well and incubated for 2 h at 37 °C, 5% CO2. The absorbance was measured with a microplate reader (FLUOstar Omega, BMG Labtech, Germany) at 450 nm.
Results and discussion
First, we have designed and synthesised 20 mer 2′-O-methyl (2′-OMe) AO sequences containing o-TINA and p-TINA on a phosphorothioate (PS) backbone based on the fully-modified 2′-OMePS AO as a positive control targeting DMD exon-23 in mdx mice myotubes (Table 1). Then, we analysed the melting temperature (Tm) of the AO sequences to the targeted complementary pre-mRNA (Table 2). Thermal stability experiments revealed that the use of p-TINA monomer at the 5′-end of the 2′-OMePS sequence led to the slight destabilisation of duplexes. On the other hand, the use of two o-TINA monomers (sequence 2o-TINA/2′-OMePS) resulted in a slight stabilisation of the duplex towards complementary pre-mRNA target. These results show that the use of both para and ortho-TINA molecules in the context of 2′-OMePS sequences and targeting RNA does not result in significantly increased thermal stability.| AO sequences | Tm, °C |
|---|---|
| a 2.0 μM of each strand in 5 mM sodium phosphate buffer, pH 7.0 containing 70 mM NaCl and 0.05 mM disodium EDTA was used. Thermal stability was measured at 260 nm vs. temperature with the temperature ramp of 1.0 °C min−1. Tm values are reported as maxima of first derivatives of melting curves with uncertainty of ±0.5 °C as determined by repetitive measurements. RNA target is: 5′-rAG GUA AGC CGA GGU UUG GCC-3′. Please see Fig. S1 (ESI) for melting curves. | |
| DNA | 65.0 |
| RNA | 77.0 |
| 2′-OMePS | 79.0 |
| 1o-TINA/2′-OMePS | 77.5 |
| 2o-TINA/2′-OMePS | 79.5 |
| 1p-TINA/2′-OMePS | 74.0 |
We then performed the in vitro stability study of the AOs using phosphodiesterase enzyme with very high 3′ → 5′ exonuclease activity. The results clearly indicated that under our conditions 2o-TINA/2′-OMePS AO was found to be relatively more stable followed by the 2′-OMePS, 1o-TINA/2′-OMePS and 1p-TINA/2′-OMePS AOs respectively (Fig. 2). In fact, the results showed the ability of TINA-modified AOs to render high nuclease stability when incorporated at the 3′-end of the sequence, whereas all other AO which doesn't have a TINA monomer at the 3′-end were comparatively less stable against phosphodiesterase enzymatic degradation.
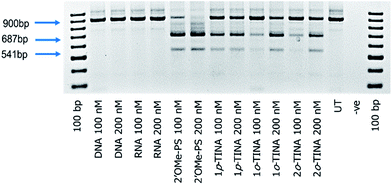
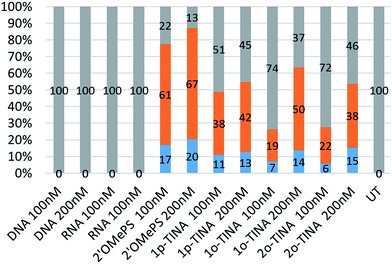
Next, we tested the feasibility of TINA-modified AOs (1p-TINA/2′-OMePS, o-TINA/2′-OMePS and 2o-TINA/2′-OMePS) in inducing exon-23 skipping. Briefly, primary mdx mouse myoblast was cultured and seeded at 2 × 104 cells per well on a 24 well plate and transfected with the AOs (Table 1) using Lipofectin as a transfection agent. Twenty-four hours after transfection, the cells were collected and isolated the RNA, and RT-PCR was performed. The products were analysed by 2% agarose gel-electrophoresis. The results clearly showed that the TINA-modified oligonucleotides successfully induced exon-23 skipping by showing product bands at 687 bp corresponding to exon-23 deletion from the full length product (900 bp) (Fig. 3). However, efficiency of exon-skipping was not as high compared to the fully-modified 2′-OMePS AO which yielded the skipped product of 687 bp in 61% at 100 nM and 67% at 200 nM concentrations (Fig. 4). AO modified with 1p-TINA/2′-OMePS showed the skipped product in 38% and 42% respectively at 100 and 200 nM concentrations. In the case of 1o-TINA/2′-OMe-PS mixmer AO, the yield of 687 bp skipped product was 50% at 200 nM although the yield at 100 nM was only 19%. Compared to 1p-TINA/2′-OMePS mixmer AO, the 1o-TINA/2′-OMePS showed superior efficacy in exon-23 skipping. This may not be surprising as 1p-TINA-modified oligonucleotides known to destabilize antiparallel duplexes (see Table 2).4,5 As we predicted an increase in the efficiency of skipping by introducing another o-TINA towards the 3′-end, we also tested the efficacy of 2o-TINA/2′-OMePS mixmer AO. But, the result showed that the yield of 687 bp skipped product was not as high compared the 1o-TINA/2′-OMePS mixmer AO. The exon skipping results are in line to the measured Tm of the corresponding duplex, however, the 2o-TINA did show notable improvement in inducing exon skipping in mdx myotubes in vitro corresponding to the observed Tm value.
Interestingly, a minor product band of 541 bp accounting for the dual exon-22/23 skipping was also observed in all of the PS-modified AOs samples in yields ranging from 6% to 20% (Fig. 4). Exon-22/23 dual skipping could shift the dystrophin gene reading frame, and therefore should be limited. Fully-modified 2′-OMePS AO showed the 541 bp exon 22/23 dual exon skipping product in high yields of 17% at 100 nM and 20% at 200 nM concentration. On the other hand, both 1p-TINA/2′-OMePS (11 & 13%) and 1o-TINA/2′-OMePS (7 & 14%) mixmer AOs showed the product in only low yields at 100 and 200 nM concentration (Fig. 4). In parallel, we also included the normal DNA and RNA AOs on a phosphodiester backbone. Unsurprisingly, both DNA and RNA AOs failed to induce any exon-23 skipping, as there was only one band detected at 900 bp corresponding to the full-length product (Fig. 3), even after increasing the AOs concentration from 100 nM to 200 nM, highlighting the significance of PS backbone to render high nucleases resistance.
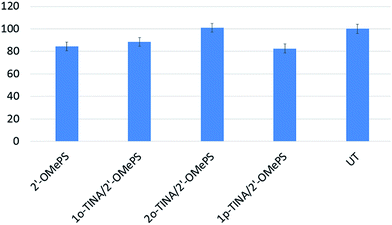
In addition, we also conducted the cell viability study to test the AOs toxicity. Overall, there was no significant difference between the treatments in terms of cell viability. However, 2o-TINA/2′-OMePS AO was found to be non-toxic under the conditions followed by 1o-TINA/2′-OMePS (88%), 2′-OMePS control (84%) and 1p-TINA/2′-OMePS (83%) respectively (Fig. 5). Although there was variation among the treatments, it was negligible when compared to the fully-modified 2′-OMePS control AO.
Conclusions
AO-mediated exon-skipping therapy is advancing as a potential therapeutic strategy in line to recent clinical trials of 2′-OMePS and phosphorodiamidate morpholino (PMO)-based AOs for the treatment of DMD. Considering this development, it is important to expand the range of chemical modifications to gain more insights on different modifications in splice modulation by induced exon-skipping. So far there are no report of using intercalating nucleic acid modifications for this purpose, and herein for the first time we investigated the applicability of TINA-modified AOs for targeted exon skipping. Introduction of both p-TINA and o-TINA together with 2′-OMePS AO did not improve the efficacy of exon-23 skipping, however, the dual exon 22/23 skipping was limited compared to the fully-modified 2′-OMePS AO counterpart. In our studies, it was clear that o-TINA was found to be a better molecule compared to the p-TINA analogue. Based on the findings, we firmly believe that the exon-skipping efficiency can be improved by a systematic screening of the positioning of TINA monomers in the sequences and also in combination with other chemistries including locked nucleic acids (LNA).14,15Acknowledgements
We thank Prof. Steve Wilton and Prof. Sue Fletcher, and their research group for providing H2K cells and their invaluable help and guidance in cell culture protocols. RNV acknowledges funding from the McCusker Charitable Foundation and the Western Australian Neuroscience Research Institute. BTL thanks the MIPS funding scheme of Murdoch University.Notes and references
- N. Dias and C. A. Stein, Mol. Cancer Ther., 2002, 1, 347–355 CAS.
- J. H. Chan, S. Lim and W. S. Wong, Clin. Exp. Pharmacol. Physiol., 2006, 33, 533–540 CrossRef CAS PubMed.
- K. E. Lundin, O. Gissberg and C. I. E. Smith, Hum. Gene Ther., 2015, 26, 475 CrossRef CAS PubMed.
- V. V. Filichev and E. B. Pedersen, J. Am. Chem. Soc., 2005, 127, 14849 CrossRef CAS PubMed.
- S. Mutsamwira, E. W. Ainscough, A. C. Partridge, P. J. Derrick and V. V. Filichev, J. Photochem. Photobiol., A, 2014, 288, 76 CrossRef CAS.
- V. V. Filichev, I. V. Astakhova, A. D. Malakhov, V. A. Korshun and E. B. Pedersen, Chem.–Eur. J., 2008, 14, 9968 CrossRef CAS PubMed.
- R. J. Fairclough, M. J. Wood and K. E. Davies, Nat. Rev. Genet., 2013, 14, 373–378 CrossRef CAS PubMed.
- C. Mitrpant, S. Fletcher and S. D. Wilton, Curr. Mol. Pharmacol., 2009, 2, 110–121 CrossRef CAS PubMed.
- T. L. Bao, R. N. Veedu, S. Fletcher and S. D. Wilton, Expert Opin. Orphan Drugs, 2016, 4, 139–152 CrossRef CAS.
- V. V. Filichev and E. B. Pedersen, J. Am. Chem. Soc., 2005, 127, 14849 CrossRef CAS PubMed.
- V. V. Filichev, I. V. Astakhova, A. D. Malakhov, V. A. Korshun and E. B. Pedersen, Chem.–Eur. J., 2008, 14, 9968 CrossRef CAS PubMed.
- J. E. Morgan, J. R. Beauchamp, C. N. Pagel, M. Peckham, P. Ataliotis, P. S. Jat, M. D. Noble, K. Farmer and T. A. Partridge, Dev. Biol., 1994, 162, 486–498 CrossRef CAS PubMed.
- C. J. Mann, K. Honeyman, A. J. Cheng, T. Ly, F. Lloyd, S. Fletcher, J. E. Morgan, T. A. Partridge and S. D. Wilton, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 42–47 CrossRef CAS.
- R. N. Veedu and J. Wengel, Chem. Biodiversity, 2010, 7, 536 CAS.
- R. N. Veedu and J. Wengel, RNA Biol., 2009, 6, 321 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Tm curve, and MS analysis of the AOs. See DOI: 10.1039/c6ra22346j |
| This journal is © The Royal Society of Chemistry 2016 |