Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Characterization of Cd36_03230p, a putative vanillin dehydrogenase from Candida dubliniensis†
Suprama
Datta
 ab,
Uday S.
Annapure
b and
David J.
Timson
ab,
Uday S.
Annapure
b and
David J.
Timson
 *ac
*ac
aSchool of Biological Sciences, Queen's University Belfast, Medical Biology Centre, 97 Lisburn Road, Belfast BT9 7BL, UK. E-mail: d.timson@brighton.ac.uk
bFood Engineering and Technology Department, Institute of Chemical Technology (ICT), Matunga, Mumbai 400 019, India
cSchool of Pharmacy and Biomolecular Sciences, University of Brighton, Huxley Building, Lewes Road, Brighton, BN2 4GJ, UK
First published on 14th October 2016
Abstract
A coding sequence (CD36-03230) from the yeast Candida dubliniensis had been previously annotated as a vanillin dehydrogenase (VDH). The corresponding protein (CD36-03230p) was recombinantly expressed in Escherichia coli and analysed. The protein is most likely a tetramer in solution as judged by crosslinking and gel filtration experiments. CD36-03230p is an active aldehyde dehydrogenase favouring cyclic and aromatic substrates. Positive cooperativity and substrate inhibition were observed with some substrates. The redox cofactor NADP+ and substrates affected the thermal stability of the protein. Interestingly, the enzyme had no detectable activity with vanillin suggesting that the annotation is incorrect. It has been previously hypothesized that a methionine residue at a key position in the active site of yeast aldehyde dehydrogenases sterically hinders cyclic substrates and restricts specificity to aliphatic aldehydes. Molecular modeling of CD36-03230p demonstrates that it has an isoleucine residue (Ile-156) at this position, further strengthening this hypothesis.
Introduction
Most mammalian aldehyde dehydrogenases (ALDHs) have a broad specificity for aliphatic aldehydes, as well as some aromatic and polycyclic aldehydes,1–3 thus rendering an important protective enzymatic function against these xenobiotics. Eubacterial ALDHs, on the other hand, exhibit relatively narrow substrate specificity depending on their natural habitat and exposure to endogenous and exogenous aldehyde reactive elements. Hence, characterization of ALDHs capable of catalyzing the oxidation of aromatic aldehydes has been well documented in bacteria.4–13Vanillin dehydrogenase (VDH), a sub-class of benzaldehyde dehydrogenases,3 is a critical enzyme for the degradation of lignin derived phenylpropanoids (such as vanillin, vanillate, caffeate, p-coumarate, cinnamate and benzaldehyde). These aromatic aldehydes, especially vanillin, are abundant as flavour and aromas in food and cosmetic industries. It is important to elucidate the structural and functional characteristics of these enzymes given their potential role in food chemistry and biotechnology. This group of enzymes has been characterized in a number of eubacterial species, including Pseudomonas fluorescens,6Pseudomonas putida KT2440,13Corynebacterium glutamicum,7Rhodococcus jostii RHA1,4Amycolatopsis sp. strain ATCC 39116,8Sphingomonas paucimobilis SYK,9 and Micrococcus sp.10 Mammalian epithelial ALDH1A and salivary ALDH3A1 typically show activity towards a wide range of aromatic aldehyde substrates (including vanillin, benzaldehyde, cinnamaldehyde, 4-hydroxynonenal).14 Plant ALDH2 family members have also been observed to show a broad aromatic substrate specificity range, but with no report of activity with vanillin.15 Two ALDHs from white rot fungus Phanerochaete chrysosporium which are translationally up-regulated with exogenous addition of vanillin are active as VDHs.16
There are reports of recombinant vanillin production in metabolically engineered baker's yeast harbouring heterologous genes,17 but no studies are available focusing on the characterization of endogenous vanillin dehydrogenase enzymes in yeast species. To date, a single protein sequence CD36-03230p (accession number: XP_002416995) from Candida dubliniensis genome has been provisionally annotated as a putative vanillin dehydrogenase (Cd36_03230p). The purpose of this study was to characterise the substrate specificity and oligomeric structure of recombinant Cd36_03230p, in order to validate (or otherwise) its putative role. The results were interpreted, in part, based on results from our previous study which described two ALDHs from Saccharomyces cerevisiae var. boulardii.18
Methods
Recombinant expression and purification of Cd36_03230p
The coding sequence for Cd36_03230 (based on accession number: XM_002416950) was synthesised following optimization of the sequence for expression in Escherichia coli (GenScript NJ, USA). The coding sequence was PCR-amplified and amplicons were inserted into the E. coli expression vector pET46 Ek/LIC (Merck-Millipore, Nottingham, UK) according to the manufacturer's instructions (note that this vector introduces bases coding for the amino acid sequence MAHHHHHHVDDDDK at the 5′ end of the coding sequence). Correct insertion into the vector was verified by PCR and by DNA sequencing (GATC, London, UK) of the insert.The expression vector was used to transform competent E. coli Rosetta™ (DE3) cells (Merck-Millipore) and colonies resulting from this transformation were used to inoculate cultures (5 ml of Luria Bertani medium (LB) supplemented with 100 μg ml−1 ampicillin and 34 μg ml−1 chloramphenicol) which were grown at 37 °C overnight (17–18 h) with orbital shaking. Each culture was then diluted into 1 l of LB (supplemented with 100 μg ml−1 ampicillin and 34 μg ml−1 chloramphenicol), grown (with orbital shaking) until A600 reached 0.6 to 1.0 (typically 5–6 h) at 30 °C, followed by a slow induction by adding 1.3 mM IPTG overnight (12–16 h) at 16 °C. These induction conditions were based on our previous experience of working with a wide variety of recombinant proteins. Cells were harvested by centrifugation (4200g for 15 min), resuspended in cell resuspension buffer (50 mM HEPES–OH, pH 7·5, 150 mM NaCl, 10% (v/v) glycerol) and stored frozen at −80 °C until the purification step.
For purification, cell suspensions were thawed, disrupted by sonication on ice (three pulses at 100 W for 30 s with 30 s gaps for cooling) and clarified by centrifugation (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 20 min, 4 °C). The supernatant was applied to a cobalt agarose column (1 ml, His-Select, Sigma, Poole, UK) which had been pre-equilibrated in buffer A (cell resuspension buffer, except 500 mM NaCl) and allowed to pass through by gravity. The column was washed with 40 ml of buffer A and the protein eluted with three 2 ml aliquots of buffer C (buffer A plus 250 mM imidazole). Protein containing fractions were identified by SDS-PAGE and dialysed overnight at 4 °C against cell resuspension buffer supplemented with 1 mM DTT. The concentration of Cd36_03230p was determined by the method of Bradford19 using BSA as a standard. The purified fractions were frozen at −80 °C in 20 μl aliquots.
000g, 20 min, 4 °C). The supernatant was applied to a cobalt agarose column (1 ml, His-Select, Sigma, Poole, UK) which had been pre-equilibrated in buffer A (cell resuspension buffer, except 500 mM NaCl) and allowed to pass through by gravity. The column was washed with 40 ml of buffer A and the protein eluted with three 2 ml aliquots of buffer C (buffer A plus 250 mM imidazole). Protein containing fractions were identified by SDS-PAGE and dialysed overnight at 4 °C against cell resuspension buffer supplemented with 1 mM DTT. The concentration of Cd36_03230p was determined by the method of Bradford19 using BSA as a standard. The purified fractions were frozen at −80 °C in 20 μl aliquots.
Bioinformatics and modeling
Multiple sequence (structure-based) alignments were carried out for Cd36_03230p with known structures of aromatic aldehyde dehydrogenases (class 3) such as benzaldehyde dehydrogenase from Pseudomonas putida (PDB ID 3LV1), Corynebacterium glutamicum (PDB ID 3R64) and a salicylaldehyde dehydrogenase from Pseudomonas putida G7 (PDB ID 4JZ6) using T-Coffee algorithm in Expresso template mode available at http://www.tcoffee.org.20–22 Human retinal dehydrogenase 1 (PDB ID 4WB9)23 and human liver mitochondrial dehydrogenase (PDB ID 1CW3)24 were also incorporated into the alignment as members of class 1 and class 2 ALDHs respectively to have an insight into their relatedness with Cd36_03230p if any. The sequence homology was evaluated using ESPript 3.0 available at http://espript.ibcp.fr.25 The phylogenetic tree was constructed using ClustalW Phylogeny (version 2.1), a web-based service available at http://www.ebi.ac.uk/Tools/msa/clustalw2/26 by neighbour-joining method.27An initial molecular model of the protein was generated using Phyre2 (ref. 28) and energy minimized using YASARA.29 A model of dimeric Cd36_03230p was generated by aligning two copies of the model to the ALDH domains of Geobacter sulfurreducens PutA (PDB 4NMB30) and saving the two monomers into a single protein structure (pdb) file. This initial, dimeric model was then subjected to a second round of energy minimization using YASARA. A tetrameric model was generated in the same way using the tetrameric structure of sheep liver class 1 aldehyde dehydrogenase (1BXS31) as the template. These models are available as ESI† to this paper.
Cross-linking
Crosslinking with bis(sulfosuccinimidyl)suberate (BS3; 50–800 μM) was carried out with 18 μM protein (diluted as required in 100 mM sodium phosphate buffer pH 7.4) in a total volume of 10 μl. Reaction mixtures were incubated at 30 °C for 30 min before addition of the crosslinker and then incubated at the same temperature for a further 35 min. Reactions were stopped by addition of an equal volume of SDS-loading buffer (120 mM Tris–HCl, pH 6.8, 4% (w/v) SDS, 20% (v/v) glycerol, 5% (w/v) bromophenol blue, 1% (w/v) DTT) and analysed by 10% SDS-PAGE.Analytical gel filtration
Cd36_03230p (200 μl of a 60 μM purified protein aliquot) was chromatographed on a Sephacryl S-300 (Pharmacia) column (total volume, Vt = 65.2 ml; void volume, V0 = 15.1 ml) at a flow rate of 1 ml min−1. The column was equilibrated and developed in buffer G (50 mM Tris–HCl, 17 mM Tris base, 150 mM sodium chloride, pH 7.4).32–34 Fractions (1 ml) were collected and analysed for protein content by measuring the absorbance at 280 nm. Standard proteins (thyroglobulin, 669 kDa; albumin, 67 kDa and chymotrypsinogen, 25 kDa) were used to calibrate the column. Their elution volumes (Ve) were used to calculate Kav according to the equation:| Kav = (Ve − V0)/(Vt − Ve) |
The Stoke's radius (Rs) was estimated from the inverse correlation of this parameter with Kav and the sedimentation coefficient (S20,w) was estimated from the molecular models using WinHydroPRO 1.00.35 The sub-unit stoichiometry (n) was then estimated using the equation:
| nM = S20,wNA(6πηRs)/(1 − v2ρ) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 Da), NA is Avogadro's number (6.023 × 1023 mol−1), η is the viscosity of the solvent (0.01 g cm−1 s−1), v2 is the partial specific volume (0.73 g cm3 g−1)36 and ρ is the density of the solvent (1.0 g cm−3). Values obtained with both the dimeric and tetrameric models of Cd36_03230p were compared in order to see which fit better to the experimental data.
100 Da), NA is Avogadro's number (6.023 × 1023 mol−1), η is the viscosity of the solvent (0.01 g cm−1 s−1), v2 is the partial specific volume (0.73 g cm3 g−1)36 and ρ is the density of the solvent (1.0 g cm−3). Values obtained with both the dimeric and tetrameric models of Cd36_03230p were compared in order to see which fit better to the experimental data.
ALDH activity measurements and enzyme kinetic analysis
The enzyme assay was performed as described previously.7,37–39 Aldehyde dehydrogenase enzyme activity was monitored at 30 °C using a ThermoScientific Multiskan™ Microplate spectrophotometer. The reactions contained 100 mM potassium phosphate buffer (pH 7.3), 0.5 mM NADP+, varied concentrations of substrates (10–1200 μM) and 0.6 μM enzyme. The long-chain (C8–C13) and phenolic aldehydes were dissolved in DMSO (1.7% (v/v), final concentration) as a solvent carrier.Steady-state kinetic data was obtained in triplicates in 96-well plates with readings taken every 5 s. The initial, linear portion of the progress curve was identified by visual inspection and fitted to linear regression to give the initial rates (v) of change in absorbance at 340 nm. These rates were converted to molar units using the extinction coefficient of NADPH (6.22 mM−1 cm−1)40 to give rates of reactions in micromolar concentration of NADH formed per second.
The kinetic parameters (kcat, K0.5 and Hill coefficient, h) were obtained by plotting the rates of reaction against substrate concentration and fitting the data to the equation below using non-linear regression as implemented in GraphPad Prism 6.0 (GraphPad Software Inc, CA). All points were weighted equally.
| v = kcat[E][S]h/(K0.5h + [S]h) |
Differential scanning fluorimetry (DSF)
Enzyme aliquots were diluted in 50 mM HEPES, pH 7.3, to a concentration of 5–7 μM to a final volume of 20 μl. Sypro Orange (10×; manufacturer's concentration definition) was as previously described.43,44 Cofactor (NADP+) and substrates were added as appropriate. Where required, substrates were initially dissolved in 100% DMSO and diluted in buffer R (50 mM HEPES–OH, pH 7.5, 150 mM NaCl, 10% v/v glycerol) as required. The concentration of DMSO never exceeded 1% (v/v).Results and discussion
Cd36_03230p is an unusual aldehyde dehydrogenase-like protein
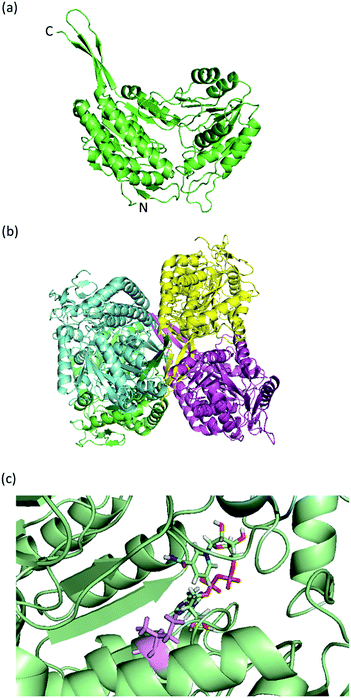
Structure-based multiple sequence alignment showed conserved residues at both NAD(P)+ binding and catalytic domains. However, the Cd36_03230p sequence has gaps and substitutions at otherwise conserved residues when aligned to class 1, class 2 and class 3 ALDHs which suggests structural and functional disparity (ESI Fig. S1a†). It also forms an independent cluster on the phylogenetic tree and exhibits clear evolutionary distance with the already known structures of class 3 benzaldehyde and salicylaldehyde dehydrogenases with verified VDH activities,7,13 besides the class 1 and 2 ALDHs (non-VDH) (ESI Fig. S1b†). Therefore, it is evident that Cd36_03230p sequence is not sufficiently similar to the salicylaldehyde dehydrogenases or benzaldehyde dehydrogenases to be classified as the member of the class 3 vanillin dehydrogenase family.45However, molecular modelling of Cd36_03230p predicted that it has a similar fold to other aldehyde dehydrogenases (Fig. 1a). The highest ranked template used in Phyre2 was PutA from G. sulfurreducens PutA (PDB 4NMB30) with a root mean squared deviation (rmsd) of 1.0 Å over 1922 equivalent atoms. The fold is also predicted to be similar to mammalian aldehyde dehydrogenases (for example, the rmsd when compared to sheep liver aldehyde dehydrogenase is 0.7 Å over 1957 equivalent atoms). The predicted structure is largely α-helical with a protruding β-sheet region at the C-terminus which is, by comparison to oligomeric aldehyde dehydrogenase structures, likely to be involved in homo-oligomer assembly. Since there are both dimeric and tetrameric aldehyde dehydrogenases known, we built both oligomeric versions in order to assist with the interpretation of gel filtration experiments (see below). Comparison with the structure of sheep liver aldehyde dehydrogenase (which was solved with NAD+ bound) enabled the prediction of the cofactor binding site. This lies near the surface of each subunit, adjacent to a cleft which is the likely aldehyde substrate binding site (Fig. 1b). Previously, we have suggested that a bulky amino acid residue (Met-177 in S. cerevisiae var. boulardii Ald6p) is partly responsible for restricting access to bulkier, cyclic aldehydes.18 In Cd36_03230p, the structurally equivalent residue is Ile-156. Therefore, we hypothesized that this smaller residue may enable Cd36_03230p to accommodate cyclic aldehydes.
Expression, purification and oligomeric structure of Cd36_03230p
Cd36_03230p could be expressed in, and purified from, E. coli Rosetta™ (DE3) cells. Typical yields were approximately 1.5 mg l−1 of bacterial cell culture (Fig. 2a). Unlike other yeast ALDHs,18 this protein did not show multiple bands resulting from oligomerisation on 10% SDS-PAGE suggesting that any oligomeric form(s) are less resistant to heat and SDS denaturation.The enzyme was able to form dimers and tetramers as demonstrated by chemical crosslinking with BS3. Resolution of the crosslinked products by 10% SDS-PAGE revealed bands corresponding primarily to a homotetramer (∼210 kDa), with some higher order oligomers (Fig. 2b). The intensity of these bands was greater following treatment with increasing concentrations of BS3. Dimeric ALDHs have previously been reported in, for example, human ALDH3 due to an extended C-terminal tail which prevents tetramerisation.46 However, our predicted structure of Cd36_03230p suggests that there is no such tail in this protein. Gel filtration chromatography was used to estimate the native molecular mass in solution and, thus the subunit stoichiometry. Sedimentation coefficients of models of the dimeric and tetrameric models of Cd36_03230p were computationally estimated as 6.3 and 10.0 S respectively in order to allow for effects due to the shape of the protein. The Stoke's radius was estimated from the gel filtration data as 5.0 nm. This yielded an estimated subunit composition of using the dimeric model of 2.5 and 4.0 using the tetrameric model. The tetrameric model is clearly a better fit to the data, suggesting the Cd36_03230p exists predominantly as a tetramer in solution. However, given that higher molecular mass species were detected by crosslinking, higher oligomeric forms may also be present.
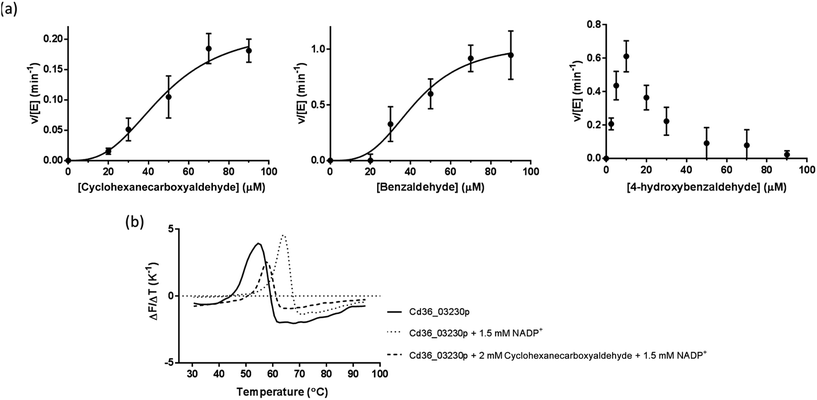
Cd36_03230p exhibits unusual kinetic patterns with cyclic and aromatic substrates
Cd36_03230p demonstrated activity with cyclohexane-carboxyaldehyde (a cyclic aliphatic aldehyde), benzaldehyde and 4-hydroxybenzaldehyde (an aromatic aldehyde), showing highest activity towards 4-hydroxy benzaldehyde judged by its kcat to K0.5 ratio (Table 1 and Fig. 3a). Interestingly, Cd36_03230p kinetics also showed substantial substrate inhibition by 4-hydroxy benzaldehyde at a substrate concentration higher than ∼10 μM. Despite attempts to fit these data to various kinetic models, it was not possible to obtain a good fit with corresponding estimates of kinetic constants for the data with this substrate. Previously, substrate inhibition of benzaldehyde dehydrogenases from Acinetobacter calcoaceticus by benzaldehyde and betaine aldehyde dehydrogenase from Staphylococcus aureus by betaine aldehyde has been reported.47,48| Substrate (CAS registry number) | k cat (min−1) | K 0.5 (μM) | k cat/K0.5 (μM−1 min) | h |
|---|---|---|---|---|
| a Although activity was observed with this compound, the kinetic data could not be fitted to Michaelis–Menten, cooperative or substrate inhibition kinetic models. b nd – no detectable turnover. Activity of Cd36_03230p was assayed using 100 mM potassium phosphate buffer (pH 7.3), 0.5 mM NADP+, varied concentrations of substrates (10–1200 μM) and 0.6 μM enzyme. Aldehydes were dissolved in 1.7% (v/v) DMSO as a solvent carrier. The final concentration of DMSO never exceeded 1% (v/v) in the assay. Values reported are those returned by non-linear fitting and are shown ± the standard errors derived from this process. | ||||
| Cyclohexanecarboxyaldehyde (2043-61-0) | 0.22 ± 0.03 | 48.0 ± 7.2 | (4.6 ± 0.2) × 10−3 | 2.9 ± 0.7 |
| Benzaldehyde (100-52-7) | 1.05 ± 0.14 | 42.8 ± 5.7 | (24.5 ± 1.5) × 10−3 | 3.2 ± 0.9 |
| 4-Hydroxybenzaldehyde (123-08-0) | a | |||
| Vanillin (121-33-5) | nd | |||
Apart from this, Cd36_03230p failed to show any activity towards aliphatic (short-chain and long-chain) and most aromatic aldehydes used in this study. These included acetaldehyde, propionaldehyde, butyraldehyde, isobutyraldehyde, valeraldehyde, hexanaldehyde, heptanaldehyde, octanaldehyde, nonanaldehyde, decyl aldehyde, undecyl aldehyde, dodecyl aldehyde, tridecyl aldehyde, crotonaldehyde, DL-glyceraldehyde and, notably, 4-hydroxy-3-methoxy benzaldehyde (vanillin).
Substrates and cofactors increases thermal stability of Cd36_03230p
Addition of NADP+ (1.5 mM) resulted in a significant increase in the “melting temperature”, Tm, of Cd36_03230p as estimated by DSF (Table 2 and Fig. 3b). This suggests that this compound binds to, and stabilizes the protein. Long-chain aliphatic and aromatic aldehyde substrates (2 mM concentration) generally reduced the thermal stability of the enzyme–NADP+ complex by ∼4 °C (Table 2). This may indicate a slightly lower stability of the ternary enzyme–NADP+–aldehyde complex, perhaps resulting from increased overall flexibility in the protein. However, given that many of the compounds which affect the thermal stability are not substrates of the enzyme (and may, therefore, not interact with the protein), it is also possible that they cause a small destabilization through their general hydrophobic or chaotropic properties rather than through interaction at a specific site.49| Substrate or cofactor | T m (°C) |
|---|---|
| a Indicates a statistically significant difference (p < 0.05). Experiments with cofactors were compared to the appropriate untreated enzyme and experiments with cofactor and aldehyde were compared to the appropriate one with cofactor only. Aldehyde substrates were measured with 2 mM aldehyde and 1.5 mM NADP+. | |
| Untreated | 53.5 ± 0.2 |
| NADP+ | 64.5 ± 0.0a |
| Propionaldehyde | 59.5 ± 0.0a |
| Valeraldehyde | 59.3 ± 0.1a |
| Octanal | 58.5 ± 0.6a |
| Decanal | 54.5 ± 0.7 |
| Tridecanaldehyde | 58.0 ± 0.3a |
| Crotonaldehyde | 59.0 ± 0.2a |
| Cyclohexanecarboxyaldehyde | 57.8 ± 0.5a |
| Benzaldehyde | 58.0 ± 0.4a |
| 4-Hydroxybenzaldehyde | 58.8 ± 0.3a |
| Vanillin | 58.0 ± 0.3a |
Conclusions
This study demonstrated that, despite being annotated as such, this enzyme has no detectable vanillin dehydrogenase activity. The data do show that the enzyme functions as an aldehyde dehydrogenase, with a strong preference for some cyclic and aromatic substrates. Our previous work suggested that a key residue in the active site influences the substrate specificity of yeast aldehyde dehydrogenases. In the case of S. cerevisiae Ald4p and Ald6p, the former has some activity to cyclic aldehyde substrates, but the latter does not.18,50,51 Ald6p has a bulky methionine residue (Met-177) which we hypothesized might sterically hinder the binding of cyclic substrates. In contrast, the structurally equivalent residue in Ald4p is Leu-196 and alteration of Met-177 in Ald6p to valine conferred some activity with cyclic aldehydes on this enzyme.18 In Cd36_03230p, the structurally equivalent residue is Ile-156. This provides further support for our hypothesis that a smaller hydrophobic residue at this position facilitates the binding of cyclic substrates. However, the lack of activity of Cd36_03230p with some cyclic substrates and all aliphatic substrates tested (in contrast with S. cerevisiae Ald4p and Ald6p which both act on aliphatic aldehydes) suggests that there are additional determinants of substrate specificity in these enzymes. Further studies are required to elucidate these. The lack of activity with vanillin suggests that Cd36_03230p's putative annotation as a vanillin dehydrogenase is incorrect and should be changed. We suggest that cyclic/aromatic aldehyde dehydrogenase would be a more appropriate annotation.Acknowledgements
This project was funded in part by the Commonwealth Scholarship Commission in the UK (CSC ref. no.: INCN-2014-46) as a split-site doctoral studentship to SD. The work was also supported by Indo-UK exchange grants from University Grants Commission, India. We thank Dr Chris CR Allen and Prof Aaron Maule (both of School of Biological Sciences, Queen's University, Belfast) for access to PCR machines used for gradient PCR and access to a qPCR machine used in the DSF assays respectively. We thank Charlotte Thomas (School of Biological Sciences, Queen's University, Belfast) for help and advice with the computational sedimentation coefficient determinations.References
- A. A. Klyosov, Biochemistry, 1996, 35, 4457–4467 CrossRef CAS PubMed.
- A. Yoshida, A. Rzhetsky, L. C. Hsu and C. Chang, Eur. J. Biochem., 1998, 251, 549–557 CAS.
- A. Marchler-Bauer, M. K. Derbyshire, N. R. Gonzales, S. Lu, F. Chitsaz, L. Y. Geer, R. C. Geer, J. He, M. Gwadz, D. I. Hurwitz, C. J. Lanczycki, F. Lu, G. H. Marchler, J. S. Song, N. Thanki, Z. Wang, R. A. Yamashita, D. Zhang, C. Zheng and S. H. Bryant, Nucleic Acids Res., 2015, 43, D222–D226 CrossRef PubMed.
- H. P. Chen, M. Chow, C. C. Liu, A. Lau, J. Liu and L. D. Eltis, Appl. Environ. Microbiol., 2012, 78, 586–588 CrossRef CAS PubMed.
- J. B. Coitinho, D. M. Costa, S. L. Guimaraes, A. M. de Goes and R. A. Nagem, Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun., 2012, 68, 93–97 CrossRef CAS PubMed.
- D. Di Gioia, F. Luziatelli, A. Negroni, A. G. Ficca, F. Fava and M. Ruzzi, J. Biotechnol., 2011, 156, 309–316 CrossRef CAS PubMed.
- W. Ding, M. Si, W. Zhang, Y. Zhang, C. Chen, L. Zhang, Z. Lu, S. Chen and X. Shen, Sci. Rep., 2015, 5, 8044 CrossRef CAS PubMed.
- C. Fleige, G. Hansen, J. Kroll and A. Steinbuchel, Appl. Environ. Microbiol., 2013, 79, 81–90 CrossRef CAS PubMed.
- E. Masai, Y. Yamamoto, T. Inoue, K. Takamura, H. Hara, D. Kasai, Y. Katayama and M. Fukuda, Biosci., Biotechnol., Biochem., 2007, 71, 2487–2492 CrossRef CAS PubMed.
- R. Mitsui, M. Hirota, T. Tsuno and M. Tanaka, FEMS Microbiol. Lett., 2010, 303, 41–47 CrossRef CAS PubMed.
- L. Tomas-Gallardo, H. Gomez-Alvarez, E. Santero and B. Floriano, Microb. Biotechnol., 2014, 7, 100–113 CrossRef CAS PubMed.
- R. Singh, V. D. Trivedi and P. S. Phale, Appl. Biochem. Biotechnol., 2014, 172, 806–819 CrossRef CAS PubMed.
- R. Plaggenborg, J. Overhage, A. Steinbuchel and H. Priefert, Appl. Biochem. Biotechnol., 2003, 61, 528–535 CAS.
- S. Solobodowska, J. Giebultowicz, R. Wolinowska and P. Wroczynski, Acta Pol. Pharm., 2012, 69, 1380–1383 CAS.
- R. Koncitikova, A. Vigouroux, M. Kopecna, T. Andree, J. Bartos, M. Sebela, S. Morera and D. Kopecny, Biochem. J., 2015, 468, 109–123 CrossRef CAS PubMed.
- T. Nakamura, H. Ichinose and H. Wariishi, Biochem. Biophys. Res. Commun., 2010, 394, 470–475 CrossRef CAS PubMed.
- E. H. Hansen, B. L. Moller, G. R. Kock, C. M. Bunner, C. Kristensen, O. R. Jensen, F. T. Okkels, C. E. Olsen, M. S. Motawia and J. Hansen, Appl. Environ. Microbiol., 2009, 75, 2765–2774 CrossRef CAS PubMed.
- S. Datta, U. S. Annapore and D. J. Timson, Appl. Biochem. Biotechnol., 2016 Search PubMed , under review.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- F. Armougom, S. Moretti, O. Poirot, S. Audic, P. Dumas, B. Schaeli, V. Keduas and C. Notredame, Nucleic Acids Res., 2006, 34, W604–W608 CrossRef CAS PubMed.
- P. Di Tommaso, S. Moretti, I. Xenarios, M. Orobitg, A. Montanyola, J. M. Chang, J. F. Taly and C. Notredame, Nucleic Acids Res., 2011, 39, W13–W17 CrossRef CAS PubMed.
- O. O'Sullivan, K. Suhre, C. Abergel, D. G. Higgins and C. Notredame, J. Mol. Biol., 2004, 340, 385–395 CrossRef PubMed.
- C. A. Morgan and T. D. Hurley, Chem.-Biol. Interact., 2015, 234, 29–37 CrossRef CAS PubMed.
- L. Ni, J. Zhou, T. D. Hurley and H. Weiner, Protein Sci., 1999, 8, 2784–2790 CrossRef CAS PubMed.
- X. Robert and P. Gouet, Nucleic Acids Res., 2014, 42, W320–W324 CrossRef CAS PubMed.
- H. McWilliam, W. Li, M. Uludag, S. Squizzato, Y. M. Park, N. Buso, A. P. Cowley and R. Lopez, Nucleic Acids Res., 2013, 41, W597–W600 CrossRef PubMed.
- N. Saitou and M. Nei, Mol. Biol. Evol., 1987, 4, 406–425 CAS.
- L. A. Kelley, S. Mezulis, C. M. Yates, M. N. Wass and M. J. E. Sternberg, Nat. Protoc., 2015, 10, 845–858 CrossRef CAS PubMed.
- E. Krieger, K. Joo, J. Lee, J. Lee, S. Raman, J. Thompson, M. Tyka, D. Baker and K. Karplus, Proteins, 2009, 77(9), 114–122 CrossRef CAS PubMed.
- H. Singh, B. W. Arentson, D. F. Becker and J. J. Tanner, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 3389–3394 CrossRef CAS PubMed.
- S. A. Moore, H. M. Baker, T. J. Blythe, K. E. Kitson, T. M. Kitson and E. N. Baker, Structure, 1998, 6, 1541–1551 CrossRef CAS PubMed.
- R. A. Durst and B. R. Staples, Clin. Chem., 1972, 18, 206–208 CAS.
- T. C. Laurent and J. Killander, J. Chromatogr. A, 1964, 14, 317–330 CrossRef CAS.
- V. L. Zinsser, E. M. Hoey, A. Trudgett and D. J. Timson, Biochimie, 2013, 95, 2182–2189 CrossRef CAS PubMed.
- A. Ortega, D. Amoros and J. Garcia de la Torre, Biophys. J., 2011, 101, 892–898 CrossRef CAS PubMed.
- H. P. Erickson, Biol. Proced. Online, 2009, 11, 32–51 CrossRef CAS PubMed.
- G. Bare, T. Swiatkowski, A. Moukil, C. Gerday and P. Thonart, Appl. Biochem. Biotechnol., 2002, 98–100, 415–428 CrossRef CAS PubMed.
- X. Peng, K. Shindo, K. Kanoh, Y. Inomata, S. K. Choi and N. Misawa, Appl. Microbiol. Biotechnol., 2005, 69, 141–150 CrossRef CAS PubMed.
- W. Yang, H. Tang, J. Ni, Q. Wu, D. Hua, F. Tao and P. Xu, PLoS One, 2013, 8, e67339 CAS.
- B. L. Horecker and A. Kornberg, J. Biol. Chem., 1948, 175, 385–390 CAS.
- P. C. Engel, in LabFax Enzymology, Bio Scientific Publishers, Oxford, UK, 1996, ch. 3 Search PubMed.
- D. J. Timson, Curr. Enzyme Inhib., 2015, 11, 12–31 CrossRef CAS.
- D. Devost and H. H. Zingg, J. Mol. Endocrinol., 2003, 31, 461–471 CrossRef CAS PubMed.
- J. Tao, H. Y. Wang and C. C. Malbon, EMBO J., 2003, 22, 6419–6429 CrossRef CAS PubMed.
- V. Vasiliou, A. Bairoch, K. F. Tipton and D. W. Nebert, Pharmacogenetics, 1999, 9, 421–434 CrossRef CAS PubMed.
- J. S. Rodriguez-Zavala and H. Weiner, Biochemistry, 2002, 41, 8229–8237 CrossRef CAS PubMed.
- R. M. Chalmers and C. A. Fewson, Biochem. J., 1989, 263, 913–919 CrossRef CAS PubMed.
- C. Chen, J. C. Joo, G. Brown, E. Stolnikova, A. S. Halavaty, A. Savchenko, W. F. Anderson and A. F. Yakunin, Appl. Environ. Microbiol., 2014, 80, 3992–4002 CrossRef PubMed.
- J. A. Cray, J. T. Russell, D. J. Timson, R. S. Singhal and J. E. Hallsworth, Environ. Microbiol., 2013, 15, 287–296 CrossRef CAS PubMed.
- M. F. Wang, C. L. Han and S. J. Yin, Chem.-Biol. Interact., 2009, 178, 36–39 CrossRef CAS PubMed.
- X. Wang, C. J. Mann, Y. Bai, L. Ni and H. Weiner, J. Bacteriol., 1998, 180, 822–830 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra22209a |
| This journal is © The Royal Society of Chemistry 2016 |