Antimicrobial activity and cytotoxicity of polyketides isolated from the mushroom Xerula sp. BCC56836†
Abstract
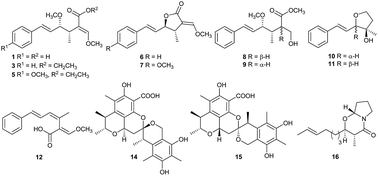
Twelve new compounds, including (−)-oudemansin A acid (1), (−)-oudemansin A ethyl ester (3) and (−)-oudemansin X ethyl ester (5), (+)-oudemansin A lactone (6) and (+)-oudemansin X lactone (7), (+)-dihydrooudemansinol (8), (−)-11-epidihydrooudemansinol (9), (+)-xeruhydrofuranol (10), (+)-9-epixeruhydrofuranol (11), xerucitrinic acids A (14) and B (15), and (2R,3R,8aR)-2-(E-hept-5-en-1-yl)-3-methyltetrahydro-2H-pyrrolo[2,1-b][1,3]oxazin-4(3H)-one (16) and one naturally new compound, strobilurin A acid (12), together with nine known compounds such as (−)-oudemansins A (2) and X (4), (3Z,5E)-3-methyl-6-phenylhexa-3,5-dien-1-ol (13), 2-(E-hept-5-en-1-yl)-3-methyl-6,7,8,8a-tetrahydro-4H-pyrrolo[2,1-b][1,3]oxazin-4-one (17), scalusamides A–C (18–20), phenol A acid (21), and dihydrocitrinone (22), were isolated from the mushroom Xerula sp. BCC56836. Their chemical structures were established based on the information from NMR spectroscopic and mass spectrometric analyses, specific rotation values, and chemical means. A plausible biosynthesis of xerucitrinic acids A (14) and B (15), the first citrinin dimers with spiro skeletons, was also proposed. In addition, the isolated compounds were evaluated for antimicrobial activity, including antimalarial, antifungal, and antibacterial activities, and their cytotoxicity against both cancerous (MCF-7, KB, NCI-H187) and non-cancerous (Vero) cells. Compounds 2, 4, and 5 possessed antimalarial activity against Plasmodium falciparum, K1 strain (IC50 1.19–13.70 μM) and compounds 3–5, 12–14, and 17 exhibited anti-Bacillus cereus activity (MIC 12.5–25.0 μg mL−1). Most isolated compounds showed low cytotoxicity against both cancerous and non-cancerous cells.

 Please wait while we load your content...
Please wait while we load your content...