Discovery of aromadendrane anologues from the marine-derived fungus Scedosporium dehoogii F41-4 by NMR-guided isolation†
Kun-Chao Huad,
Meng-Yang Xua,
Hou-Jin Lib,
Jie Yuanc,
Ge Tangc,
Jun Xu a,
De-Po Yangad and
Wen-Jian Lan*ad
a,
De-Po Yangad and
Wen-Jian Lan*ad
aSchool of Pharmaceutical Sciences, Sun Yat-sen University, Guangzhou 510006, China. E-mail: lanwj@mail.sysu.edu.cn
bSchool of Chemistry and Chemical Engineering, Sun Yat-sen University, Guangzhou 510275, China
cZhongshan School of Medicine, Sun Yat-sen University, Guangzhou 510080, China
dGuangdong Technology Research Center for Advanced Chinese Medicine, Guangzhou 510006, China
First published on 29th September 2016
Abstract
Eight closely biogenetically-related aromadendrane-type sesquiterpenoids, six of them new, scedogiines A–F (1–3, 6–8), together with a new polyketide scedogiine G (9) were discovered from the culture broth of the marine-derived fungus Scedosporium dehoogii F41-4 by NMR-guided isolation. Their structures were determined from extensive NMR and HR-ESI-MS data. The absolute configurations of the new sesquiterpenoids were elucidated by ECD calculation. Plausible biogenetic pathways of the aromadendrane analogues were postulated.
1. Introduction
Aromadendranes possess a carbon skeleton of a ger-dimethylcyclopropane fused to a hydroazulene, and have been discovered in a variety of organisms, such as plants, liverworts, microorganisms, corals and sponges.1 This group of sesquiterpenoinds has attracted much attention for their structural uniqueness and diversity as well as their wide range of biological properties including cytotoxic,2 antifungal,3 antibacterial,4 antiviral,5 neuroprotective,6 antifouling,7 and other activities.8,9 Synthetic chemists have developed various routes to construct these molecular scaffolds.10–13 Microbiologists have utilized fungal strains as cell factories to produce these sesquiterpenoids.14,15 This family of compounds displayed highly shielded protons in the range of δH −0.2–0.8 with a doublet of doublets (dd) or a doublet of doublets of doublets (ddd) splitting pattern with at least a coupling constant of about 9.6. Previously, we isolated a pair of aromadendrane diastereoisomers from a marine-derived fungus Pseudallescheria boydii F19-1.16 In our continuous screening of the fungi associated with invertebrates such as soft corals, starfishes, and sponges to obtain new and bioactive metabolites, we have isolated and purified a fungal strain authenticated as Scedosporium dehoogii F41-4 from the sponge Phyllospongia foliascens collected in the Hainan Sanya National Coral Reef Reserve, China. Scedosporium dehoogii is a member of Pseudallescheria/Scedosporium species complex, opportunistic fungal pathogens which can cause potentially fatal invasive infections in immunocompromised patients and immunocompetent individuals.17 A literature search revealed the research about the fungus Scedosporium dehoogii focusing on the identification and distribution17–19 and none of any report on the chemical investigation up to date. The ethyl acetate extraction of the culture broth in a small-scale cultivation displayed several characteristic resonance signals of cylopropanes in the range of δH −0.2–0.8 in the 1H NMR spectrum, implying the possible presence of aromadendrane sesquiterpenoids. Therefore, we determined to investigate the secondary metabolites of the fungus Scedosporium dehoogii F41-4. By tracking characteristic 1H NMR signals in the high shield region, six new aromadendrane-type sesquiterpenoids scedogiines A–F (1–3, 6–8), two known analogues pseuboydones A, and B (4, 5), were isolated, along with a new polyketide scedogiine G (9). Here, we report the isolation, structure determination and proposed biosynthetic pathways of these compounds.2. Results and discussion
2.1 Structural elucidation
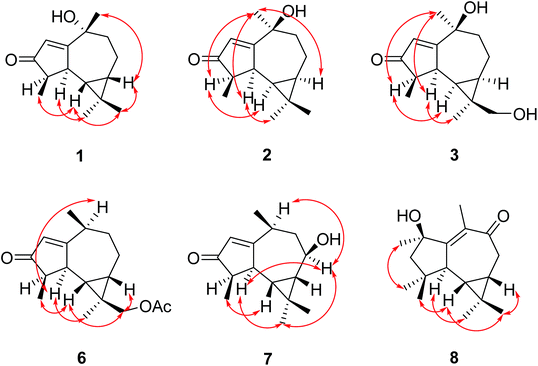
Scedogiine A (1) showed a molecular ion peak at m/z 235.1684 [M + H]+ in the HRESIMS spectrum, corresponding to a molecular formula of C15H22O2, indicating five degrees of unsaturation. The 13C NMR and DEPT spectra showed four methyls, two methylenes, five methines, and four quaternary carbons. The 1H NMR spectrum displayed two characteristic signals of a cyclopropane ring at δH −0.04 (dd, 10.0, 10.0), and 0.64 (ddd, 10.0, 12.0, 5.6) for H-6 and H-7. A sp2 methine at δH/δC 5.97(s)/128.4, a sp2 quaternary carbon at δC 181.8 and a carbonyl group at δC 211.7 revealed the existence of an α,β-unsaturated ketone, which was confirmed by the HMBC correlations from H-2 to C-1 and C-3. Apart from these signals as described above, the remaining two sites of unsaturation are attributed to two additional rings in 1. The COSY correlations of H3-15/H-4, H-4/H-5, H-5/H-6, H-6/H-7, H-7/H-8, H-8/H-9 revealed the heptyl moiety (C15–C4–C9). Further analysis of the HMBC correlations of H-4/C-3, H3-15/C-3, H-5/C-3, H-5/C-1, H-5/C-1, H-9/C-10, H-9/C-1 deduced a five- and seven-membered fused ring system. The singlet methyl-14 at δH 1.58 was connected to the quaternary carbon C-10 from the HMBC interaction between H3-14 and C-10. The geminal methyls were attached to the quaternary carbon C-11 position based on the HMBC correlations of H3-12/C-11 and H3-13/C-11. The remaining hydroxyl group was ambiguously linked to C-10 matching well with the chemical shift at δ 83.1 for the oxygenated quaternary carbon. It was supposed that cooperative influence of the electron-donating effect of two methyls, α-orientation of H-6 and steric effect of the C-10 substituents caused the highly shielded signal of H-6 at δ −0.04 (Fig. 1).The relative configuration was determined by the NOESY experiment. The NOE cross peaks of H3-15 with H-6, H3-12 with H-6 and H-7, H3-14 with H-7 implied that Me-15, H-6, H-7, Me-12, and Me-14 were oriented to the same side, the NOE interaction between H-5 and H3-13 suggested H-5 and Me-13 were placed on the other side. The absolute stereochemistry of compound 1 was achieved by ECD computed method. The ECD curve for (4R,5S,6S,7S,10R)-1 matched well with the experimental one. Therefore, the absolute configuration of compound 1 was determined as 4R, 5S, 6S, 7S, 10R.
The molecular formula of scedogiine B (2) was deduced to be C15H22O2, which is the same molecular formula as 1, from the molecular ion peak at m/z 235.1688 [M + H]+ in the HRESIMS spectrum. Careful inspection of 1D NMR spectra of compound 2 revealed that the splitting patterns of protons and types of carbons are identical to those in 1. The only difference is the chemical shift between them, which suggested that compound 2 is a stereoisomer of 1. The planar structure of 2 was further confirmed by the 1H-1H COSY and HMBC spectra data. The cross-peaks of H-4 with H-6 and H3-13, H-5 with H3-13 and H3-14, and H-7 with H3-14 inferred that H-4, H-5, H-6, H-7, Me-13, and Me-14 were oriented on the same side, whereas Me-15, Me-12, and OH-10 were located on the other side. Finally, the calculated ECD spectrum for (4R,5S,6R,7R,10S)-2 had an excellent fit with the experimental one. Therefore, the absolute configuration of 2 was assigned as 4R, 5S, 6R, 7R, 10S.
The HRESIMS spectrum of scedogiine C (3) gave a molecular ion peak at m/z 273.1455 [M + Na]+, indicating a molecular formula of C15H22O3, one more oxygen atom than that of 2. The NMR spectra of compound 3 are similar to those of compound 2. The significant difference is that the hydroxylated methylene at C-12 in 3 in place of methyl-12 in 2. The NOESY spectrum of 3 also displayed the similar relative configuration with that of 2. By ECD calculation, the absolute stereochemistry of compound 3 was assigned as 4R, 5S, 6R, 7R, 10S, 11R.
Scedogiine D (6) demonstrated a molecular ion peak at m/z 299.1615 [M + Na]+. From its HRESIMS spectrum, a molecular formula of C17H24O3 was inferred, requiring six degrees of unsaturation. Its NMR data pattern resembles that of compound 4. The main difference is that an acetoxy group in 6 instead of a hydroxyl group in 4, in accordance with the molecular formula of 6. The NOE cross peaks of H3-15 with H-6, H2-12 with H-6 and H-7, H-5 with H-10 and H3-13 suggested that Me-15, H-6, H-7, CH2-12, and Me-14 were placed on the same orientation, whereas H-4 and Me-13 were located on the opposite direction. The absolute configuration was unambiguously elucidated as 4R, 5S, 6S, 7S, 10S, 11R by comparing the computed ECD spectrum with the experimental CD curve.
From the molecular ion peak at m/z 235.1689 [M + H]+ in the HRESIMS spectrum, scedogiine E (7) was found to have the same molecular formula of C15H22O2 as compound 1. The NMR data are similar to those of 1. The difference is the hydroxyl group substituted at the C-8 position at δH/δC 3.74/69.4 in 7 instead of the C-10 position at δC 83.1 in 1. This was supported by the 1H-1H correlations between H-7 and H-8, and between H-8 and H2-9. In the NOESY spectrum, the interactions of H3-15 with H-6, H3-12 with H-6 and H-7, H-5 with H3-13, H3-13 with H-8, and H-8 with H-10 indicated that Me-15, H-6, H-7, Me-12, and Me-14 were syn-oriented, while H-4, H-5, Me-13, H-8, and H-10 were on the opposite side. The absolute configuration was assigned as 4R, 5S, 6R, 7S, 8R, 10S by comparing the calculated ECD spectrum with the experimental one (Fig. 2).
The molecular formula of scedogiine F (8) was established as C15H22O2 on the analysis of the HRESIMS peak at m/z 235.1682 [M + H]+, implying five degrees of unsaturation. Two highly shielded proton signals at δH 1.00 (dd, 9.6, 11.2), and 0.78 (ddd, 9.6, 5.6, 11.6) inferred the presence of a cyclopropane ring. The 13C and DEPT NMR spectra displayed signals at δC 133.7, 164.1, and 201.9 of an α,β-unsaturated ketone. Above spectroscopic data indicated compound 8 also possessed an aromadendrane skeleton as compounds 1–7. The 1H-1H COSY couplings between H-2 and H2-3 and HMBC correlations from H3-14 to C-10, C-1, and C-9, from H2-8 to C-9, from H-2, H-3, and H-5 to C-1 undoubtedly determined the substituted positions of the hydroxyl and α,β-unsaturated ketone groups. The NOE correlations of Me-12 with H-6 and H-7, H-6 with Me-15, H-2 with H-4, H-5 with Me-13 revealed that Me-15, Me-12, H-6, and H-7 were β-oriented, while H-2, H-4, H-5 and Me-13 were α-positioned. The calculated ECD spectrum for (2R,4S,5R,6S,7S)-8 showed an excellent fit with the experimental one. Therefore, the absolute configuration of compound 8 was determined as 2R, 4S, 5R, 6S, 7S (Fig. 3).
The molecular formula of compound 9 was established as C20H34O3 based on the molecular ion peak at m/z 321.2429 [M − H]− in the HRESIMS spectrum, requiring four double bond equivalents. The 13C NMR and DEPT spectra revealed five sp2 quaternary carbon resonances at δC 166.2, 164.9, 162.2, 106.2, 98.1 due to the typical α-pyrone carbons in the range of unsaturated carbon, accounting for the sites of unsaturation. Thus the remaining moiety was aliphatic. The 1H NMR spectrum showed two olefinic methyl singlets substituted on the α-pyrone ring, four aliphatic methyl doublets, and one aliphatic methyl triplet. Additionally, the 1H NMR spectrum also showed a series of methylene and/or methine protons with close chemical shifts, which resulted in the indistinctive 1H-1H COSY couplings. However, a 4,6,8-trimethyldecanyl side chain was clearly inferred based on the HMBC couplings of H3-18 with C-7 and C-8, H3-19 with C-9, C-8 and C-10, H3-20 with C-11, C-10, and C-12, H3-21 with C-13, C-12 and C-14, H3-15 with C-14 and C-13. The connection of the alkyl chain with the α-pyrone ring was confirmed from the HMBC interactions from H-8, and H3-18 to C-6, and the weak correlation from H-7 to C-6. Unfortunately, the sample was changed into a mixture with very intricate signals, which prevented us to further stereochemistry elucidation (Fig. 4).
The chemical structures of two known sesquiterpenoids were determined as pseuboydones A (4), and B (5) by comparing their spectroscopic data with the literature values (Fig. 5).
2.2 The proposed biosynthetic pathways
Obviously, compounds 1–8 are derived from the melavonic acid pathway. Therefore, the biosynthetic pathways of these compounds were proposed as the following. The process initiated with the conversion of acetyl-coenzyme A (CoA) to the isopentenyl pyrophosphate (IPP), which generated the sesquiterpenoid precursor farnesyl pyrophosphate (FPP) catalyzed by various prenyltransferases.20 The elimination of the pyrophosphate group resulted in the formation of the carbocation B, which performed dehydration, followed by cyclization to form stereoisomers D1 and D2. Then dehydrogenation of D1 at C-1/C-2 and C-1/C-10 positions and D2 at C-1/C-2 position furnished the intermediates E1-1, E1-2, and E2 respectively, which were oxygenated to yield compound 8 and the intermediates F1 and F2 separately. Further oxygenation of F1 at C-10, C-12, or C-8 positions and oxygenation of F2 at C-10 or C-12 positions individually generated compounds 1, 4, 7, 2, and 5 individually. Finally, esterization of 4 and further oxygenation of 3 at C-12 position to obtain the products 6 and 3 separately. Thus it can be seen that the cyclization of carbon cation B can simultaneously occur the configuration conversion of C-6/C-7. Additionally, the generation of compounds 2 and 3 indicated that oxygenation of C-10 can also accompany the configuration alteration.3. Experimental
3.1 General procedures
1D and 2D NMR spectra were generated on Bruker Avance II 400 spectrometers (Bruker BioSpin AG, Industriestrasse 26, Fällanden, Switzerland). The chemical shifts are corresponding to the residual solvent signals (CDCl3: δH 7.26 and δC 77.0, acetone-d6: δH 2.05, CD3OD δH 3.31). The low- and high resolution ESI-MS spectra were obtained with Thermo LCQ DECA XP liquid chromatography-mass spectrometry and Thermo Fisher LTQ Orbitrap Elite High Resolution liquid chromatography-mass spectrometry respectively. Optical rotations were measured on an Anton paar MCP500 polarimeter. CD spectra were performed on a JASCO J-810 circular dichroism spectrometer (JASCO International Co. Ltd., Hachioji, Tokyo, Japan). UV spectra were recorded on a Shimadzu UV-VIS-NIR spectrophotometer (Shimadzu Corporation, Nakagyo-ku, Kyoto, Japan). IR spectra were acquired on a Fourier transform infrared (FT-IR) spectrophotometer (PerkinElmer Frontier) with an Ever-Glomid/near-IR source. Preparative HPLC was carried on a Shimadzu LC-20AT HPLC pump (Shimadzu Corporation, Nakagyo-ku, Kyoto, Japan) outfitted with an SPD-20A dual λ absorbance detector (Shimadzu Corporation, Nakagyo-ku, Kyoto, Japan) and a Shim-pack PRC-ODS HPLC column I(250 × 20 mm i.d., 5 μm, Shimadzu Corporation, Nakagyo-ku, Kyoto, Japan), a Capcell pak C-18 HPLC column II(250 × 20 mm i.d., 5 μm, Shiseido Company, Limited, Nihonbashi, Japan), using a flow rate of 4.0 ml min−1 at room temperature. Silica gel (SiO2, 100–200 mesh and 200–300 mesh) for column chromatography (c.c) was purchased from Qingdao Marine Chemical Factory, Qingdao, China. Sephadex LH-20 (GE healthcare Bio-Sciences Corp., Piscataway, USA) was obtained from green herbs, Beijing, China.3.2 Fungal material
The marine fungal Scedosporium dehoogii F41-4 was isolated from the inner tissue of the sponge Phyllospongia foliascens collected from Hainan Sanya National Coral Reef Reserve, P. R. China. This fungal strain was maintained in 15% glycerol aqueous solution at −80 °C. A voucher specimen was deposited in the School of Pharmaceutical Sciences, Sun Yat-sen University, Guangzhou, P. R. China. Analysis of the ITS rDNA (GenBank KP132700) by BLAST database screening provided a 99.9% match to Scedosporium dehoogii.3.3 Culture, extraction, and isolation
This fungal strain was cultured in 500 ml Erlenmeyer flasks containing 250 ml sterile liquid nutrient media, which consisted of glucose 10 g, peptone 5 g, yeast extract 2 g, CaCO3 1 g and sea water 1 L. The flasks were incubated at 28 °C for 40 days. Then the fermentation broth was carried out pretreatment to sweep impurity through the filtration of cheesecloth. The fermentation broth was extracted three times with EtOAc at room temperature, and the organic solvent was evaporated in vacuum to afford the crude extract (10.32 g). The crude extract was chromatographed on silica gel (200–300 mesh) with a gradient solvent system of petroleum ether–EtOAc–MeOH (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0–0
0–0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, v/v/v) to give twelve major fractions (1–12). Fraction 2 (0.82 g) was subjected to a Sephadex LH-20 column and eluted with MeOH to gain three fractions (2.1, 2.2, 2.3). Compound 1 (1.30 mg) was obtained from fraction 2.3 by preparative reversed phase HPLC (MeOH–H2O, 58
10, v/v/v) to give twelve major fractions (1–12). Fraction 2 (0.82 g) was subjected to a Sephadex LH-20 column and eluted with MeOH to gain three fractions (2.1, 2.2, 2.3). Compound 1 (1.30 mg) was obtained from fraction 2.3 by preparative reversed phase HPLC (MeOH–H2O, 58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 42, v/v, tR = 108 min, column II). Fraction 3 (1.52 g) was chromatographed on SiO2 with a gradient solvent system of petroleum ether-EtOAc (10
42, v/v, tR = 108 min, column II). Fraction 3 (1.52 g) was chromatographed on SiO2 with a gradient solvent system of petroleum ether-EtOAc (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–1
1–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, v/v, 100–200 mesh), yielding four fractions (3.1, 3.2, 3.3, 3.4). The compounds 8 (1.10 mg) and 9 (5.20 mg) were obtained from fraction 3.2 by the further purification using Sephadex LH-20 and preparative reversed phase HPLC (MeOH–H2O, 80
3, v/v, 100–200 mesh), yielding four fractions (3.1, 3.2, 3.3, 3.4). The compounds 8 (1.10 mg) and 9 (5.20 mg) were obtained from fraction 3.2 by the further purification using Sephadex LH-20 and preparative reversed phase HPLC (MeOH–H2O, 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, v/v, tR8 = 37.5 min, tR9 = 41 min, column II). Fraction 3.3 was subjected to preparative reversed phased HPLC (MeOH–H2O, 56
20, v/v, tR8 = 37.5 min, tR9 = 41 min, column II). Fraction 3.3 was subjected to preparative reversed phased HPLC (MeOH–H2O, 56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 44, v/v, tR = 20 min, column I) to furnish compound 3 (0.90 mg), fraction 3.4 was subjected to Sephadex LH-20 and preparative reversed phased HPLC (MeOH–H2O, 75
44, v/v, tR = 20 min, column I) to furnish compound 3 (0.90 mg), fraction 3.4 was subjected to Sephadex LH-20 and preparative reversed phased HPLC (MeOH–H2O, 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25, v/v, tR = 40.5 min, column I) to yield compound 6 (1.33 mg). Fraction 4 (0.56 g) was subjected to column chromatography over ODS (MeOH–H2O, 9
25, v/v, tR = 40.5 min, column I) to yield compound 6 (1.33 mg). Fraction 4 (0.56 g) was subjected to column chromatography over ODS (MeOH–H2O, 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–1
1–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, v/v) to yield six major subfractions (4.1, 4.2, 4.3, 4.4, 4.5, 4.6), fraction 4.3 were further purified via preparative reversed phase HPLC (MeOH–H2O, 70
9, v/v) to yield six major subfractions (4.1, 4.2, 4.3, 4.4, 4.5, 4.6), fraction 4.3 were further purified via preparative reversed phase HPLC (MeOH–H2O, 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, v/v, tR4 = 24 min, tR5 = 26.5 min, column I) to give compound 4 (1.53 mg) and 5 (1.40 mg). Fraction 6 (0.52 g) was subjected to chromatographed on Sephadex LH-20 preparative reversed phase HPLC (MeOH–H2O, 68
30, v/v, tR4 = 24 min, tR5 = 26.5 min, column I) to give compound 4 (1.53 mg) and 5 (1.40 mg). Fraction 6 (0.52 g) was subjected to chromatographed on Sephadex LH-20 preparative reversed phase HPLC (MeOH–H2O, 68![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 32, v/v, tR = 20.5 min, column I) to give compound 7 (1.08 mg). Fraction 9 (1.15 g) was subjected to Sephadex LH-20 column using MeOH to afford four fractions, the second of which was chromatographed by preparative reversed phase HPLC (MeOH–H2O, 58
32, v/v, tR = 20.5 min, column I) to give compound 7 (1.08 mg). Fraction 9 (1.15 g) was subjected to Sephadex LH-20 column using MeOH to afford four fractions, the second of which was chromatographed by preparative reversed phase HPLC (MeOH–H2O, 58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 42, v/v, tR = 18 min, column I) to obtain compound 2 (0.95 mg).
42, v/v, tR = 18 min, column I) to obtain compound 2 (0.95 mg).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε): 227 (3.78). IR (KBr)νmax: 3384, 2972, 2926, 2869, 1705, 1600, 1456, 1404, 1377, 1262, 1090, 1051, 881, 802 cm−1. 1H and 13C NMR data see Tables 1 and 2. HRESIMS m/z 235.1684 [M + H]+ (calcd for C15H23O2, 235.1693).
ε): 227 (3.78). IR (KBr)νmax: 3384, 2972, 2926, 2869, 1705, 1600, 1456, 1404, 1377, 1262, 1090, 1051, 881, 802 cm−1. 1H and 13C NMR data see Tables 1 and 2. HRESIMS m/z 235.1684 [M + H]+ (calcd for C15H23O2, 235.1693).
| No. | 1 | 2 | 3 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|
| 1 | 181.8, C | 185.8, C | 188.6, C | 187.0, C | 185.9, C | 164.1, C | |
| 2 | 128.4, CH | 124.6, CH | 124.5, CH | 125.7, CH | 126.0, CH | 75.1, CH | 166.2, C |
| 3 | 211.7, C | 214.1, C | 212.3, C | 211.1, C | 211.1, C | 42.5, CH2 | 98.1, C |
| 4 | 47.4, CH | 50.7, CH | 50.6, CH | 46.9, CH | 46.9, CH | 32.3, CH | 164.9, C |
| 5 | 41.7, CH | 45.6, CH | 45.4, CH | 44.5, CH | 45.9, CH | 45.4, CH | 106.2, C |
| 6 | 29.7, CH | 32.2, CH | 28.4, CH | 25.8, CH | 28.9, CH | 31.8, CH | 162.2, C |
| 7 | 27.9, CH | 28.7, CH | 25.3, CH | 25.6, CH | 35.1, CH | 22.9, CH | 32.2, CH |
| 8 | 18.0, CH2 | 18.6, CH2 | 19.7, CH2 | 23.1, CH2 | 69.4, CH | 42.3, CH2 | 41.5, CH2 |
| 9 | 38.6, CH2 | 41.1, CH2 | 42.2, CH2 | 35.7, CH2 | 45.7, CH2 | 201.9, C | 28.0, CH |
| 10 | 83.1, C | 71.5, C | 75.0, C | 40.3, CH | 36.5, CH | 133.9, C | 46.2, CH2 |
| 11 | 20.8, C | 20.9. C | 26.7, C | 24.0, C | 20.4, C | 26.0, C | 27.3, CH |
| 12 | 28.3, CH3 | 28.7, CH3 | 72.5, CH2 | 73.8, CH2 | 28.1, CH3 | 28.3, CH3 | 44.6, CH2 |
| 13 | 15.9, CH3 | 15.9, CH3 | 11.7, CH3 | 12.1, CH3 | 15.5, CH3 | 16.0, CH3 | 31.8, CH |
| 14 | 23.7, CH3 | 30.0, CH3 | 28.0, CH3 | 19.9, CH3 | 19.5, CH3 | 14.5, CH3 | 30.6, CH2 |
| 15 | 10.0, CH3 | 17.8, CH3 | 18.1, CH3 | 10.0, CH3 | 9.8, CH3 | 16.5, CH3 | 11.6, CH3 |
| 16 | 171.3, C | 8.7, CH3 | |||||
| 17 | 21.0, CH3 | 9.8, CH3 | |||||
| 18 | 19.3, CH3 | ||||||
| 19 | 20.7, CH3 | ||||||
| 20 | 19.9, CH3 | ||||||
| 21 | 19.1, CH3 |
| No. | 1 | 2 | 3 |
|---|---|---|---|
| 1 | |||
| 2 | 5.97 (s) | 5.84 (s) | 6.09 (s) |
| 3 | |||
| 4 | 2.66 (qd, 7.2, 6.4) | 2.30 (q, 7.2) | 2.31 (q, 7.6) |
| 5 | 2.94 (dd, 10.0, 6.4) | 2.56 (d, 9.6) | 2.10 (d, 9.6) |
| 6 | −0.04 (dd, 10.0, 10.0) | 0.22 (dd, 9.6, 9.6) | 0.45 (dd, 9.6, 9.6) |
| 7 | 0.64 (ddd, 10.0, 12.0, 5.6) | 0.66 (ddd, 9.6, 10.8, 6.4) | 0.88 (ddd, 9.6, 5.6, 11.6) |
| 8 | 1.75 (dddd, 14.8, 5.6, 5.6, 1.2) | 1.79 (ddd, 14.4, 6.4, 6.4) | 1.10 (m) |
| 1.48 (dddd, 14.8, 12.0, 12.0, 1.2) | 1.55 (ddd, 14.4, 11.6, 11.6) | 2.00 (m) | |
| 9 | 2.11 (ddd, 14.8, 5.6, 1.2) | 1.98 (dd, 12.4, 6.4) | 1.83 (dd, 12.0, 12.0) |
| 1.65 (ddd, 14.8, 12.0, 1.2) | 1.69 (dd, 12.4, 12.4) | 2.00 (m) | |
| 10 | |||
| 11 | |||
| 12 | 1.03 (s) | 1.04 (s) | 3.34 (s) |
| 13 | 1.09 (s) | 1.14 (s) | 1.21 (s) |
| 14 | 1.58 (s) | 1.56 (s) | 1.40 (s) |
| 15 | 1.15 (d, 7.2) | 1.14 (d, 7.2) | 1.15 (d, 7.6) |
| 16 | 1.94 (brs) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε): 229 (3.85). IR (KBr)νmax: 3403, 2959, 2926, 2869, 1690, 1601, 1568, 1457, 1374, 1265, 1174, 1150, 869, 810, 681 cm−1. 1H and 13C NMR data see Tables 1 and 2 HRESIMS m/z 235.1688 [M + H]+ (calcd for C15H23O2, 235.1693).
ε): 229 (3.85). IR (KBr)νmax: 3403, 2959, 2926, 2869, 1690, 1601, 1568, 1457, 1374, 1265, 1174, 1150, 869, 810, 681 cm−1. 1H and 13C NMR data see Tables 1 and 2 HRESIMS m/z 235.1688 [M + H]+ (calcd for C15H23O2, 235.1693).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε): 231 (4.03). IR (KBr)νmax: 3378, 2972, 2925, 2870, 1688, 1600, 1459, 1409, 1375, 1172, 1086, 1049, 1031, 881, 870 cm−1. 1H and 13C NMR data see Tables 1 and 2 HRESIMS m/z 273.1455 [M + Na]+ (calcd for C15H22O3Na, 273.1461).
ε): 231 (4.03). IR (KBr)νmax: 3378, 2972, 2925, 2870, 1688, 1600, 1459, 1409, 1375, 1172, 1086, 1049, 1031, 881, 870 cm−1. 1H and 13C NMR data see Tables 1 and 2 HRESIMS m/z 273.1455 [M + Na]+ (calcd for C15H22O3Na, 273.1461).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε): 230 (3.84). IR (KBr)νmax: 3418, 2956, 2927, 2871, 1738, 1705, 1601, 1457, 1379, 1246, 1229, 1048, 1028, 982 cm−1. 1H and 13C NMR data see Tables 1 and 3. HRESIMS m/z 299.1615 [M + Na]+ (calcd for C17H24O3Na, 299.1618).
ε): 230 (3.84). IR (KBr)νmax: 3418, 2956, 2927, 2871, 1738, 1705, 1601, 1457, 1379, 1246, 1229, 1048, 1028, 982 cm−1. 1H and 13C NMR data see Tables 1 and 3. HRESIMS m/z 299.1615 [M + Na]+ (calcd for C17H24O3Na, 299.1618).
| No. | 6 | 7 | 8 | 9 |
|---|---|---|---|---|
| 1 | ||||
| 2 | 5.84 (d, 1.6) | 5.84 (d, 1.6) | 4.80 (dd, 6.8, 6.8) | |
| 3 | 2.25 (ddd, 12.8, 6.8, 6.8) | |||
| 1.62 (ddd, 12.8, 9.6, 7.6) | ||||
| 4 | 2.63 (qd, 6.8, 6.8) | 2.58 (qd, 6.8, 6.8) | 2.14 (m) | |
| 5 | 2.62 (dd, 6.8, 10.4) | 2.49 (dd, 10.0, 6.8) | 2.62 (dd, 9.6, 9.6) | |
| 6 | 0.29 (dd, 9.6, 9.6) | 0.22 (dd, 10.0, 10.0) | 1.00 (dd, 9.6, 11.2) | |
| 7 | 0.89 (ddd, 9.6, 11.2, 5.6) | 0.76 (dd, 10.0, 10.0) | 0.78 (ddd, 9.6, 5.6, 11.6) | 3.01 (m) |
| 8 | 2.06 (m) | 3.74 (ddd, 10.0, 10.0, 1.2) | 2.87 (dd, 14.8, 5.6) | 1.83 (ddd, 14.0, 10.0, 4.0) |
| 1.24 (m) | 2.33 (dd, 14.8, 11.6) | 1.14 (dd, 4.0, 4.0) | ||
| 9 | 1.99 (m) | 2.06 (ddd, 12.4, 4.8, 1.2) | 1.32 (ddddd, 6.8) | |
| 1.37 (ddd, 12.4, 12.4, 12.4) | 1.72 (dd, 12.4, 12.4, 10.0) | |||
| 10 | 2.34 (m) | 2.46 (m) | 1.11 (dddd, 6.8, 6.8, 6.8, 6.8) | |
| 0.91 (dd, 6.8, 6.8) | ||||
| 11 | 1.55 (ddddd, 6.8) | |||
| 12 | 3.89 (d, 11.2) | 1.10 (s) | 1.08 (s) | 0.95 (dd, 6.8) |
| 3.74 (d, 11.2) | ||||
| 13 | 1.18 (s) | 1.19 (s) | 1.17 (s) | 1.35 (ddddd, 6.8) |
| 14 | 1.24 (d, 7.6) | 1.26 (d, 6.8) | 1.95 (dd, 1.2, 1.2) | 1.09 (dddd, 6.8, 6.8, 6.8, 6.8) |
| 1.22 (dd, 7.6, 6.0) | ||||
| 15 | 1.14 (d, 7.6) | 1.11 (d, 6.8) | 1.10 (d, 7.2) | 0.84 (t, 7.2) |
| 16 | 2.00 (s) | |||
| 17 | 2.04 (s) | 2.00 (s) | ||
| 18 | 1.17 (d, 6.8) | |||
| 19 | 0.83 (d, 7.2) | |||
| 20 | 0.73 (d, 6.4) | |||
| 21 | 0.79 (d, 6.4) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε): 224.5 (3.99), 281 (3.27). IR (KBr)νmax: 3359, 2959, 2926, 2871, 2856, 1693, 1601, 1571, 1458, 1414, 1377, 1310, 1263, 1177, 1089, 1049, 881 cm−1. 1H and 13C NMR data see Tables 1 and 3 HRESIMS m/z 235.1689 [M + H]+ (calcd for C15H23O2, 235.1693).
ε): 224.5 (3.99), 281 (3.27). IR (KBr)νmax: 3359, 2959, 2926, 2871, 2856, 1693, 1601, 1571, 1458, 1414, 1377, 1310, 1263, 1177, 1089, 1049, 881 cm−1. 1H and 13C NMR data see Tables 1 and 3 HRESIMS m/z 235.1689 [M + H]+ (calcd for C15H23O2, 235.1693).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε): 243 (3.63). IR (KBr)νmax: 3393, 2957, 2926, 2871, 2856, 1711, 1645, 1567, 1455, 1415, 1377, 1303, 1091, 1052, 989, 979, 883 cm−1. 1H and 13C NMR data see Tables 1 and 3 HRESIMS m/z 235.1682 [M + H]+ (calcd for C15H23O2, 235.1693).
ε): 243 (3.63). IR (KBr)νmax: 3393, 2957, 2926, 2871, 2856, 1711, 1645, 1567, 1455, 1415, 1377, 1303, 1091, 1052, 989, 979, 883 cm−1. 1H and 13C NMR data see Tables 1 and 3 HRESIMS m/z 235.1682 [M + H]+ (calcd for C15H23O2, 235.1693).3.4 Computational methods
The absolute configurations of compounds 1–3 and 6–8 were determined by quantum chemical calculation of ECD spectra using Gaussian 09 software.21 Conformational searches were performed by the MOE software using the MMFF94 molecular mechanics force field.22 Within a 2 kcal mol−1 energy window. After the conformational search, 3 conformations with the lowest energy were found for 1, 2, 7 and 8, 4 for 3, 6 for 6 (Fig. S1†). The molecule lowest energy conformers were geometrically optimized using the DFT method at the B3LYP/6-31+G(d) level in acetonitrile. The ECD spectra for the optimized conformers were calculated using the TDDFT method at the PBE1PBE/6-311++G (d, p) level in acetonitrile.23,24 The overall theoretical ECD spectra were obtained according to the Boltzmann weighting of each conformer. The ECD spectra were generated by the SpecDis 1.6 software25 using a Gaussian band shape with a 0.3 eV exponential half-width from dipole-length dipolar and rotational strengths.4. Conclusion
Although numerous novel natural products have been obtained from a large number of marine-derived fungi, actually the discovering rate of the new entitles is still not high. Developing technologies for efficient dereplication of known compounds is still one of the major challenges for chemical investigation of fungal origin. In this paper we illustrated an example to effectively increase the discovery rate of new compounds by tracking the diagnostic resonance signals.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This project is financially supported by Guangdong Provincial Science and Technology Research Program (No. 2013B021100010, 2013B021100012, 2014A020217004, 2015A020216007, and 2016A020222004), Guangzhou Science and Technology Research Program (No. 2014J4100059), the Fundamental Research Funds for the Central Universities (No. 15ykpy05 and 14yksh01), and instrumental analysis fund of Sun Yat-sen University (No. 0504015080000).Notes and references
- M. J. Duran-Pena, J. M. B. Ares, J. R. Hanson, I. G. Collado and R. Hernandez-Galan, Nat. Prod. Rep., 2015, 32, 1236–1248 RSC.
- A. E. Nugroho, R. Sugiura, T. Momota, Y. Hirasawa, C. P. Wong, T. Kaneda, A. H. A. Hadi and H. Morita, J. Nat. Med., 2015, 69, 411–415 CrossRef CAS PubMed.
- I. C. Moreira, J. H. G. Lago, M. C. M. Young and N. F. Roque, J. Braz. Chem. Soc., 2003, 14, 828–831 CrossRef CAS.
- M. Gilabert, K. Marcinkevicius, S. Andujar, M. Schiavone, M. E. Arena and A. Bardon, Phytomedicine, 2015, 22, 77–85 CrossRef CAS PubMed.
- N. De Tommasi, C. Pizza, C. Conti, N. Osi and M. L. Stein, J. Nat. Prod., 1990, 53, 830–835 CrossRef CAS.
- S. K. S. Amoah, D. Vecchia, M. T. B. Pedrini, G. L. Carnhelutti, A. E. Goncalves, D. A. dos Santos, M. W. Biavatti and M. M. de Souza, Eur. J. Pharmacol., 2015, 769, 195–202 CrossRef CAS PubMed.
- H. Hirota, Y. Tomono and N. Fusetani, Tetrahedron, 1996, 52, 2359–2368 CrossRef CAS.
- T. D. Hubert and D. F. Wiemer, Phytochemistry, 1985, 24, 1197–1198 CrossRef CAS.
- Y. Asakawa, M. Toyota, T. Takemoto, I. Kubo and K. Nakanishi, Phytochemistry, 1980, 19, 2147–2154 CrossRef CAS.
- J. Carreras, M. Livendahl, P. R. McGonigal and A. M. Echavarren, Angew. Chem., Int. Ed., 2014, 53, 4896–4899 CrossRef CAS PubMed.
- M. Calancea, S. Carret and D. Jean-Pierre, Eur. J. Org. Chem., 2009, 19, 3134–3137 CrossRef.
- G. L. Lange and A. Merica, Tetrahedron Lett., 1999, 40, 7897–7900 CrossRef CAS.
- S. L. Gwaltney and K. J. Shea, Tetrahedron Lett., 1996, 37, 949–952 CrossRef CAS.
- D. O. Collins, P. L. D. Ruddock, J. C. de Grasse, W. F. Reynolds and P. B. Reese, Phytochemistry, 2002, 59, 479–488 CrossRef CAS PubMed.
- K. P. McCook, A. R. M. Chen, W. F. Reynolds and P. B. Reese, Phytochemistry, 2012, 82, 61–66 CrossRef CAS PubMed.
- W. Lan, K. Wang, M. Xu, J. Zhang, C. Lam, G. Zhong, J. Xu, D. Yang, H. Li and L. Wang, RSC Adv., 2016, 6, 76206–76213 RSC.
- A. Harun, F. Gilgado, S. C. A. Chen and W. Meyer, Med. Mycol., 2010, 48(suppl. 1), 70–76 CrossRef PubMed.
- (a) F. Gilgado, J. Cano, J. Gene, D. A. Sutton and J. Guarro, J. Clin. Microbiol., 2008, 46, 766–771 CrossRef PubMed; (b) E. Sitterle, S. Giraud, J. Leto, J. P. Bouchara, A. Rougeron, F. Morio, B. Dauphin, C. Angebault, G. Quesne, J. L. Beretti, N. Hassouni, X. Nassif and M. E. Bougnoux, Clin. Microbiol. Infect., 2014, 9, 929–935 CrossRef PubMed.
- A. Rougeron, G. Schuliar, J. Leto, E. Sitterle, D. Landry, M. E. Bougnoux, A. Kobi, J. P. Bouchara and S. Giraud, Environ. Microbiol., 2015, 17(4), 1039–1048 CrossRef CAS PubMed.
- D. J. McGarvey and R. Croteau, Plant Cell, 1995, 7, 1015–1026 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci and G. A. Petersson, Gaussian 09, Revision B.01, Gaussian, Pittsburgh, PA, 2009 Search PubMed.
- A. Schäfer, H. Horn and R. Ahlrichs, J. Chem. Phys., 1992, 97, 2571–2577 CrossRef.
- J. P. Perdew, K. Burke and M. Ernzerhof, Phys. Rev. Lett., 1997, 78, 1396 CrossRef CAS.
- A. D. Rabuck and G. E. Scuseria, Chem. Phys. Lett., 1999, 309, 450–456 CrossRef CAS.
- T. Bruhn, A. Schaumloffel, Y. Hemberger and G. Bringmann, Chirality, 2013, 25, 243–249 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: The NMR spectra data of compounds 1–9. See DOI: 10.1039/c6ra21142a |
| This journal is © The Royal Society of Chemistry 2016 |