Luminescence properties of a Zn(II) supramolecular framework: easily tunable optical properties by variation of the alkyl substitution of (E)-N-(pyridine-2-ylethylidyne)arylamine ligands†
Yu-Wei Dong,
Rui-Qing Fan*,
Wei Chen,
Hui-Jie Zhang,
Yang Song,
Xi Du,
Ping Wang,
Li-Guo Wei and
Yu-Lin Yang*
MIIT Key Laboratory of Critical Materials Technology for New Energy Conversion and Storage, School of Chemistry and Chemical Engineering, Department of Chemistry, Harbin Institute of Technology, Harbin 150001, P. R. China. E-mail: fanruiqing@hit.edu.cn; ylyang@hit.edu.cn; Fax: +86-451-86413710
First published on 15th November 2016
Abstract
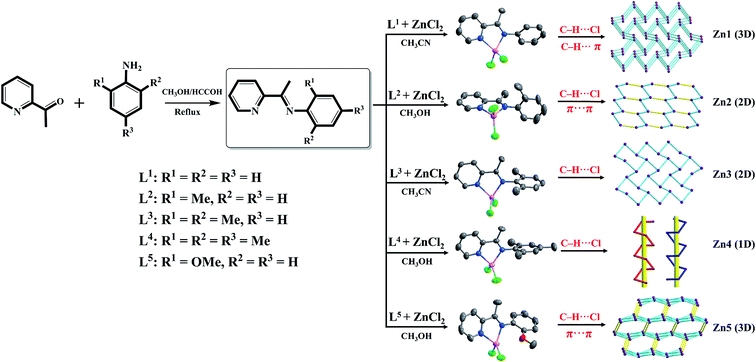
A series of different alkyl substituted of Zn(II) complexes, [ZnL1Cl2] (Zn1), [ZnL2Cl2] (Zn2), [ZnL3Cl2] (Zn3), [ZnL4Cl2] (Zn4), [ZnL5Cl2] (Zn5) ((E)-N-(pyridine-2-yl)(CMe![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NPhR), where R = H, L1; 2-CH3, L2; 2,6-(CH3)2, L3; 2,4,6-(CH3)3, L4; 2-OCH3, L5) have been synthesized and characterized by single crystal X-ray diffraction, 1H NMR, FT-IR, and EA. The X-ray diffraction analyses revealed that although are all constructed by C–H⋯Cl/π hydrogen bonds and π⋯π interactions, the dimensions of these supramolecular frameworks complexes Zn1–Zn5 are quite different. Complexes Zn1 and Zn5 feature 3D 5-connected {46·64} and {44·66} topology structures, respectively, while complexes Zn2 and Zn3 feature 2D supramolecular layer with {63} topology structures. Complex Zn4 exhibits two different one-dimensional helix-shaped chains. Obviously, these results show that steric hindrance has a great impact on the final structures of the supramolecular architectures. Based on these varied structures caused by different alkyl substitutions, the emission maximum wavelengths of complexes Zn1–Zn5 can be tuned in a large range of 400–514 nm. The λem shift in the red direction after the substitution of alkyl is attributed to the HOMO–LUMO energy gap of complexes being effectively decreased due to the electron-donating ability of alkyl. These results are confirmed by the density functional theory calculations.
NPhR), where R = H, L1; 2-CH3, L2; 2,6-(CH3)2, L3; 2,4,6-(CH3)3, L4; 2-OCH3, L5) have been synthesized and characterized by single crystal X-ray diffraction, 1H NMR, FT-IR, and EA. The X-ray diffraction analyses revealed that although are all constructed by C–H⋯Cl/π hydrogen bonds and π⋯π interactions, the dimensions of these supramolecular frameworks complexes Zn1–Zn5 are quite different. Complexes Zn1 and Zn5 feature 3D 5-connected {46·64} and {44·66} topology structures, respectively, while complexes Zn2 and Zn3 feature 2D supramolecular layer with {63} topology structures. Complex Zn4 exhibits two different one-dimensional helix-shaped chains. Obviously, these results show that steric hindrance has a great impact on the final structures of the supramolecular architectures. Based on these varied structures caused by different alkyl substitutions, the emission maximum wavelengths of complexes Zn1–Zn5 can be tuned in a large range of 400–514 nm. The λem shift in the red direction after the substitution of alkyl is attributed to the HOMO–LUMO energy gap of complexes being effectively decreased due to the electron-donating ability of alkyl. These results are confirmed by the density functional theory calculations.
Introduction
The concept of “crystal engineering”1,2 was revolutionized with the participation of coordination polymers and metallosupramolecular architectures.3–6 The latter is not only designed and developed for their diverse structural aesthetics but also for a variety of potential applications in catalysis, separation, sensors, adsorption, luminescent materials, magnetism, etc.7–11 The glue for the inorganic supramolecular synthesis is the metal–ligand coordinate-covalent bond whose strength and directionality have been thoroughly exploited for engineering various molecular building blocks into diverse supramolecular architectures.12–14 The dimensionality of the resulting coordination network can be increased by noncovalent interactions, such as hydrogen bonds, π⋯π, C–H⋯π, aromatic ring⋯halogen, and/or other van der Waals interactions.15–20 Schiff base ligands are frequently used in coordination chemistry due to their significant ability to form stable complexes with metalions.21,22 Zinc Schiff base complexes with luminescent properties can be used in applications involving fabrication of novel materials and as probes in biological systems.23 Controllable tuning of fluorescent properties of zinc complexes based on ligand design is always a challenge for chemists. From the viewpoint of molecular and crystal engineering, the critical criterion for ligand design is how to embed noncovalent interaction groups to control the molecular structure and the supramolecular framework.The type and position of substituents are the main factors to affect the supramolecular frameworks and luminescent properties. In line with the above discussion, five (E)-N-(pyridine-2-ylethylidyne)arylamine ligands, L1–L5, Scheme 1, carrying different alkyl substitutions in the phenyl ring have been employed for the synthesis of Zn(II) complexes. Five Zn(II) complexes, namely Zn1–Zn5, were prepared by the reactions of corresponding ligand and zinc chloride, which display luminescence ranging from deep blue to bluish green. The optical properties of all the complexes were investigated by UV-vis absorption and luminescence spectroscopy in solution and in the solid state. The electronic effects of the substituents on the ligand were also determined as important factors for the modulation of the luminescence wavelength. In this report we will also carry out density functional theory (DFT) calculations in both the ground and excited state of the complexes for understanding of the electronic structures.
Experimental section
Materials and instrumentation
All reagents and solvents were purchased from commercial sources and were used without further purification. Elemental analyses were carried out on a Perkin-Elmer 2400 automatic analyzer. FT-IR spectra data (4000–400 cm−1) were collected by a Nicolet impact 410 FT-IR spectrometer. 1H NMR spectra were obtained using a Bruker Avance-400 MHz spectrometer with Si(CH3)4 as internal standard. The thermal analyses were performed on a ZRY-2P thermogravimetric analysis under a flow of air from room temperature to 700 °C. Powder X-ray diffraction (PXRD) patterns were recorded in the 2θ range of 5–50° using Cu-Kα radiation by Shimadzu XRD-6000 X-ray Diffractometer. A Perkin-Elmer Lambda 35 spectrometer was used to measure the UV-vis absorption spectra of ligands and complexes. The emission luminescence and lifetime properties were recorded with Edinburgh FLS 920 fluorescence spectrometer.X-ray crystallography
Suitable crystals of complexes Zn1–Zn5 were selected and mounted on a Rigaku R-AXIS RAPID IP diffractometer. Diffraction data were collected using graphite-monochromatized Mo Kα radiation (λ = 0.71073 Å). The structures were solved with direct methods24 and refined with full-matrix least-squares on F2. All the hydrogen atoms were constrained in geometric positions to their parent atoms and non-hydrogen atoms were refined anisotropically. The detailed crystal structure refinement data are given in Table 1, selected bond lengths and angles are listed in Tables S1 and S2.† The CCDC numbers are 1494305–1494308, 1477759 for Zn1–Zn5, respectively.†| Zn1 | Zn2 | Zn3 | Zn4 | Zn5 | |
|---|---|---|---|---|---|
| a R1 = ∑||Fo| − |Fc||/∑|Fo|; wR2 = [∑[w(Fo2 − Fc2)2]/∑[w(Fo2)2]]1/2. | |||||
| CCDC no. | 1494305 | 1494306 | 1494307 | 1494308 | 1477759 |
| Formula | C13H12Cl2N2Zn | C14H14Cl2N2Zn | C15H16Cl2N2Zn | C16H18Cl2N2Zn | C14H14Cl2N2OZn |
| Mr | 332.52 | 346.56 | 360.57 | 374.61 | 362.54 |
| Cryst. syst. | Monoclinic | Triclinic | Monoclinic | Monoclinic | Monoclinic |
| Space group | P21/c | P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P21/n | C2/c | P21/n |
| a [Å] | 9.558(5) | 8.108(5) | 9.095(4) | 32.804(14) | 11.311(3) |
| b [Å] | 10.911(5) | 13.089(8) | 11.742(5) | 13.390(5) | 9.796(3) |
| c [Å] | 13.640(6) | 15.690(9) | 15.094(7) | 16.187(6) | 14.305(4) |
| α [°] | 90 | 71.786(7) | 90 | 90 | 90 |
| β [°] | 92.856(4) | 76.105(7) | 91.716(10) | 96.815(8) | 106.827(3) |
| γ [°] | 90 | 84.537(7) | 90 | 90 | 90 |
| Volume [Å3] | 1420.82(12) | 1535.1(16) | 1611.48(12) | 7060(5) | 1517.1(8) |
| Z | 4 | 4 | 4 | 16 | 4 |
| Dc [g cm−3] | 1.554 | 1.499 | 1.486 | 1.410 | 1.587 |
| μ [mm−1] | 2.087 | 1.935 | 1.846 | 1.689 | 1.966 |
| F(000) | 672 | 704 | 736 | 3071 | 736 |
| θ range [°] | 3.25–27.56 | 1.40–26.81 | 3.11–27.58 | 1.25–25.00 | 2.03–26.93 |
| h range | −12 ≤ h ≤ 12 | −10 ≤ h ≤ 10 | −11 ≤ h ≤ 11 | −38 ≤ h ≤ 38 | −13 ≤ h ≤ 14 |
| k range | −12 ≤ k ≤ 14 | −16 ≤ k ≤ 15 | −15 ≤ k ≤ 15 | −15 ≤ k ≤ 15 | −12 ≤ k ≤ 12 |
| l range | −6 ≤ l ≤ 17 | −19 ≤ l ≤ 15 | −19 ≤ l ≤ 19 | −19 ≤ l ≤ 19 | −18 ≤ l ≤ 17 |
| Data/restraints/params | 3241/0/163 | 5771/30/346 | 3722/0/181 | 6209/0/379 | 3041/0/181 |
| GOF | 0.808 | 1.034 | 1.003 | 1.063 | 1.023 |
| R1, wR2[I > 2σ(I)]a | 0.0390, 0.1119 | 0.0382, 0.0944 | 0.0405, 0.1224 | 0.0405, 0.1454 | 0.0336, 0.0807 |
| R1, wR2[all data]a | 0.0583, 0.1309 | 0.0681, 0.1084 | 0.0541, 0.1350 | 0.0515, 0.1580 | 0.0479, 0.0873 |
| Δρmax, Δρmin [e Å−3] | 0.346, −0.418 | 0.448, −0.530 | 0.601, −0.341 | 1.435, −0.997 | 0.379, −0.370 |
Computational details
To gain insights into the photophysical properties of complexes Zn1–Zn5, and the energy transfer process from the free ligand to the complexes, their energy levels based on density functional theory (DFT) theoretical calculations were performed. The geometries obtained from X-ray diffraction analyses were fully optimized first in conjunction with the acetonitrile solvent model based on B3LYP/6-31G basis set. The theoretical investigation of geometry optimization was performed with the Gaussian 09 program package.25 Density functional theory (DFT) was calculated at Beck's three-parameter hybrid exchange functional26 and Lee, and Yang and Parr correlation functional27 B3LYP/6-31G(d). The spin density distributions were visualized using Gaussview 5.0.8.General synthesis of zinc(II) complexes
The zinc(II) complexes were synthesized by dissolving metal salts ZnCl2 and 1 molar equivalence of the respective Schiff-base ligands in anhydrous solutions (acetonitrile or methanol). The yellow solutions were refluxed for 12 h and subsequently filtered. X-ray quality single crystals of five Zn(II) complexes were grown from slow evaporation of their solutions (Scheme 1). In addition, the detailed IR and 1H NMR data for complexes Zn1–Zn5 are listed in the Table S3.†![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N 1623, νZn–N 446.
N 1623, νZn–N 446.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N 1632, νZn–N 438.
N 1632, νZn–N 438.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N 1631, νZn–N 439.
N 1631, νZn–N 439.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N 1623, νZn–N 433.
N 1623, νZn–N 433.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N 1620, νZn–N 437.
N 1620, νZn–N 437.Results and discussion
Synthesis and spectral characterization
The five imine ligands (E)-N-(pyridine-2-yl)(CMe![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NPhR) (where R = H, L1; 2-CH3, L2; 2,6-(CH3)2, L3; 2,4,6-(CH3)3, L4; 2-OCH3, L5) were readily prepared in a one-step procedure by condensation of 2-acetylpyridine with the corresponding monamine in a 1
NPhR) (where R = H, L1; 2-CH3, L2; 2,6-(CH3)2, L3; 2,4,6-(CH3)3, L4; 2-OCH3, L5) were readily prepared in a one-step procedure by condensation of 2-acetylpyridine with the corresponding monamine in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio in anhydrous methanol/formic acid solution (see Scheme 1). Yields for the five imine ligands varied from 81 to 92%. Using the self-assembled method, these ligands were reacted with ZnCl2 to obtain a series of complexes, namely, Zn1–Zn5. X-ray quality single crystals of five Zn(II) complexes were grown from slow evaporation of methanol or acetonitrile solutions and readily obtained in good yields within the range of 53–65%. The details of the synthesis are given in the Experimental section. We confirm that all the complexes are stable in the solid state under exposure to air. The complexes are soluble in common organic solvents, such as chloroform, dimethylsulfoxide and acetonitrile.
1 molar ratio in anhydrous methanol/formic acid solution (see Scheme 1). Yields for the five imine ligands varied from 81 to 92%. Using the self-assembled method, these ligands were reacted with ZnCl2 to obtain a series of complexes, namely, Zn1–Zn5. X-ray quality single crystals of five Zn(II) complexes were grown from slow evaporation of methanol or acetonitrile solutions and readily obtained in good yields within the range of 53–65%. The details of the synthesis are given in the Experimental section. We confirm that all the complexes are stable in the solid state under exposure to air. The complexes are soluble in common organic solvents, such as chloroform, dimethylsulfoxide and acetonitrile.
The infrared spectra of these five Zn(II) complexes (see Fig. S1 and S2†) are similar to that of the corresponding ligand and the IR assignments of selected diagnostic bands are given in the Experimental section. In the IR spectra of ligands L1–L5, the existence of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bonds is clearly demonstrated by the presence of strong characteristic C
N bonds is clearly demonstrated by the presence of strong characteristic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N peaks in a range of 1625–1657 cm−1. These bands undergo negative shifts of 2–34 cm−1 in the complexes, which may be attributed to the coordination of the nitrogen atom of the imine to the metal ion.28,29 This is further confirmed by the presence of a ν(M − N) vibration in the region 433–446 cm−1 in all the complexes.30 The 1H NMR spectra of the imine ligands L1–L5 and the corresponding Zn(II) complexes were recorded in CDCl3 at room temperature (see Fig. S3 and S4†). In the 1H NMR spectra, the chemical shift values of the protons in the complexes are slightly different from those observed for the non-coordinated ligand. Of particular note is the Py-H6 resonances of the complexes Zn1–Zn5, which are moved down-field due to coordination between the pyridine nitrogen and the zinc(II) center. Meanwhile, the resonance peaks at 3.87 (L5) and 4.08 (Zn5) ppm are assigned to –OCH3 protons.
N peaks in a range of 1625–1657 cm−1. These bands undergo negative shifts of 2–34 cm−1 in the complexes, which may be attributed to the coordination of the nitrogen atom of the imine to the metal ion.28,29 This is further confirmed by the presence of a ν(M − N) vibration in the region 433–446 cm−1 in all the complexes.30 The 1H NMR spectra of the imine ligands L1–L5 and the corresponding Zn(II) complexes were recorded in CDCl3 at room temperature (see Fig. S3 and S4†). In the 1H NMR spectra, the chemical shift values of the protons in the complexes are slightly different from those observed for the non-coordinated ligand. Of particular note is the Py-H6 resonances of the complexes Zn1–Zn5, which are moved down-field due to coordination between the pyridine nitrogen and the zinc(II) center. Meanwhile, the resonance peaks at 3.87 (L5) and 4.08 (Zn5) ppm are assigned to –OCH3 protons.
The thermal stability of the five complexes was investigated by the TGA technique (see Fig. S5, ESI†). The TGA curves of complexes Zn1–Zn3 are similar and thermally decomposed in two steps. The first weight loss of 20.63% at around 271 °C, due to the removal of 2HCl molecules (calc. 21.32%) in Zn1, and 25.22% at around 291 °C, due to the removal of 2HCl + (–CH3) molecules (calc. 24.78%) in Zn2, and 27.70% at around 310 °C, due to the removal of 2HCl + 2(–CH3) molecules (calc. 27.98%) in Zn3, respectively. The following decompositions of Zn1–Zn3 in the temperature range of 381–649 °C. For Zn4 and Zn5, there are three continuous weight loss steps among them. The first weight loss can be attributed 2HCl molecules, which is approximately 19.44% for Zn4 and 19.13% for Zn5 (calc. 18.92% for Zn4 and 19.55% for Zn5). The following weight losses of 11.03% for Zn4 are attributed to the loss of three –CH3 (calc. 12.01%) and 14.21% for Zn5 can ascribe to the release of one –OCH3 and one –CH3 (calc. 12.69%). These complexes finally transformed to the final residual of the ZnO phase (calc. 24.47%, 23.48%, 22.57%, 18.92%, and 22.44% for Zn1–Zn5, respectively), which are in agreement with the experimental results (found 24.97%, 23.54%, 22.81%, 19.13%, and 22.81% for Zn1–Zn5, respectively).
We performed powder X-ray diffraction patterns on complexes Zn1–Zn5 to check the purity of the bulk products. From Fig. S6,† we can see that all major peak positions of the measured patterns are in good agreement with those simulated. Furthermore, the differences in intensity may be due to the preferred orientation of the crystal products. These observations are consistent with the crystal structure (see below). To further understand the structures of these complexes, single crystals were obtained and analyzed by single crystal X-ray diffraction. The crystallographic data for these complexes Zn1–Zn5 are listed in Table 1.
Description of the structures
| Complex | Addison parameter (τ4) | Dihedral anglea (degree) | D–H⋯A | D–H (Å) | H⋯A (Å) | D⋯A (Å) | D–H⋯A (degree) | Supramolecular dimension |
|---|---|---|---|---|---|---|---|---|
| a Between the pyridyl ring and phenyl ring.b Cg1 = phenyl ring.c Cg2 = pyridyl ring. | ||||||||
| Zn1 | 0.890 | 60.441 | C7–H7C⋯Cl2 | 0.960 | 2.678 | 3.384 | 130.782(2) | 3D |
| C12–H12A⋯Cl1 | 0.930 | 2.896 | 3.570 | 130.422(2) | ||||
| C3–H3A⋯πCg1b | 3.036 | |||||||
| Zn2 | 0.899, 0.891 | 83.403, 85.146 | C2–H2A⋯Cl4 | 0.930 | 2.889 | 3.563 | 133.430(4) | 2D |
| C3–H3A⋯Cl1 | 0.930 | 2.897 | 3.702 | 145.695(5) | ||||
| C17–H17A⋯Cl3 | 0.930 | 2.890 | 3.685 | 144.210(5) | ||||
| πCg1⋯πCg1 | 3.741, 3.851 | |||||||
| Zn3 | 0.876 | 80.205 | C4–H4A⋯Cl1 | 0.930 | 2.908 | 3.561 | 128.428(2) | 2D |
| C7–H7A⋯Cl1 | 0.960 | 2.916 | 3.656 | 134.690(2) | ||||
| Zn4 | 0.901, 0.879 | 87.987, 84.936 | C3–H3A⋯Cl3 | 0.930 | 2.895 | 3.735 | 150.869(2) | 1D |
| C19–H19A⋯Cl1 | 0.930 | 2.934 | 3.716 | 142.632(2) | ||||
| C30–H30B⋯Cl2 | 0.960 | 2.889 | 3.689 | 141.490(3) | ||||
| Zn5 | 0.791 | 49.456 | C4–H4A⋯Cl2 | 0.930 | 2.741 | 3.648 | 165.258(2) | 3D |
| C10–H10A⋯Cl2 | 0.930 | 2.875 | 3.746 | 156.511(1) | ||||
| πCg2⋯πCg2c | ||||||||
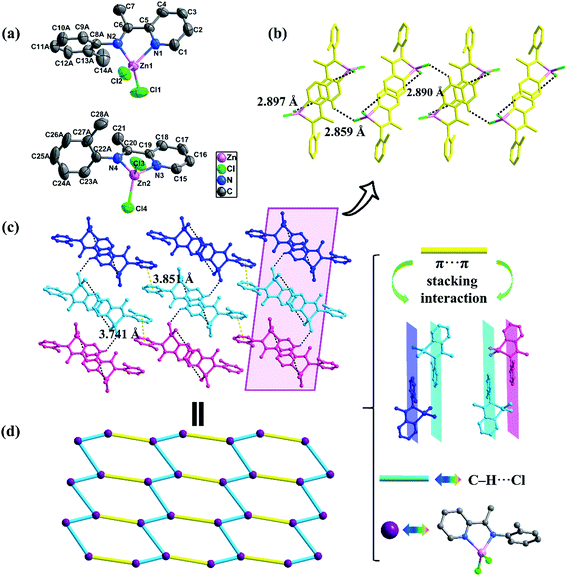
![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) space group. The asymmetric unit consists of two crystallographically and conformationally independent molecules as shown in Fig. 2a. In Zn2, two zinc atoms are bonded to four nitrogen atoms with similar distances varying from 2.055(3) to 2.073(3) Å (Table S2†). The coordination geometry of Zn2+ are distorted tetrahedron, with four-coordinated geometry index τ4 = 0.899 (Zn1) and 0.891 (Zn2). The bond angles around the Zn(II) ion are in the range of 79.32(1)–118.15(9)°. The dihedral angles between the pyridyl ring and the phenyl ring in the two molecules are 83.403° and 85.146°, respectively (Table 2), which are seriously twisty compared with the complex Zn1. From Fig. 2b, the independent units are linked through C2–H2A⋯Cl4, C3–H3A⋯Cl1 and C17–H17A⋯Cl3 hydrogen bonds interactions to generate a one-dimensional “wave-like” chain. The H2A⋯Cl4, H3A⋯Cl1 and H17A⋯Cl3 distances are 2.859, 2.890 and 2.897 Å, respectively. In Zn2, except the C–H⋯Cl hydrogen bonds interactions, the intermolecular π⋯π stacking interactions play important roles in assembling the two-dimensional structures. As shown in Fig. 2c, a 2D layer can be constructed by interlayer πphenyl⋯πphenyl (3.741 and 3.851 Å) stacking interactions. Topology analysis shows that the overall network of Zn2 features a 2D hcb topology structure with a Schläfli symbol of {63} by denoting the mononuclear units as 3-connected nodes and the C–H⋯Cl and π⋯π interactions as the aqua and yellow linkers, respectively (Fig. 2d).
space group. The asymmetric unit consists of two crystallographically and conformationally independent molecules as shown in Fig. 2a. In Zn2, two zinc atoms are bonded to four nitrogen atoms with similar distances varying from 2.055(3) to 2.073(3) Å (Table S2†). The coordination geometry of Zn2+ are distorted tetrahedron, with four-coordinated geometry index τ4 = 0.899 (Zn1) and 0.891 (Zn2). The bond angles around the Zn(II) ion are in the range of 79.32(1)–118.15(9)°. The dihedral angles between the pyridyl ring and the phenyl ring in the two molecules are 83.403° and 85.146°, respectively (Table 2), which are seriously twisty compared with the complex Zn1. From Fig. 2b, the independent units are linked through C2–H2A⋯Cl4, C3–H3A⋯Cl1 and C17–H17A⋯Cl3 hydrogen bonds interactions to generate a one-dimensional “wave-like” chain. The H2A⋯Cl4, H3A⋯Cl1 and H17A⋯Cl3 distances are 2.859, 2.890 and 2.897 Å, respectively. In Zn2, except the C–H⋯Cl hydrogen bonds interactions, the intermolecular π⋯π stacking interactions play important roles in assembling the two-dimensional structures. As shown in Fig. 2c, a 2D layer can be constructed by interlayer πphenyl⋯πphenyl (3.741 and 3.851 Å) stacking interactions. Topology analysis shows that the overall network of Zn2 features a 2D hcb topology structure with a Schläfli symbol of {63} by denoting the mononuclear units as 3-connected nodes and the C–H⋯Cl and π⋯π interactions as the aqua and yellow linkers, respectively (Fig. 2d).
Photophysical studies
| Complex | Absorptiona | Photoluminescence in acetonitrile | Photoluminescence in the solid state | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| λabs/nm (ε/M−1 cm−1) | λem/nm | FWHM/nm | τ/μs | ΦPLb | CIE (x, y) | λem/nm | FWHM/nm | τ/μs | CIE (x, y) | |
| a Measured in CH3CN solutions (∼1 × 10−5 M).b Take quinine sulfate in CH3CN as reference (∼1 × 10−5 M, ΦPL = 0.546). | ||||||||||
| L1 | 269 (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 895), 323 (1357) 895), 323 (1357) |
393 | 80.15 | 6.10 | 0.020 | 0.19, 0.13 | 458 | 147.11 | 7.93 | 0.18, 0.25 |
| Zn1 | 280 (40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 427), 323 (18 427), 323 (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 193) 193) |
400 | 101.40 | 9.15 | 0.095 | 0.17, 0.12 | 472 | 134.85 | 9.90 | 0.22, 0.29 |
| L2 | 269 (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 688), 329 (2216) 688), 329 (2216) |
397 | 134.20 | 8.10 | 0.025 | 0.18, 0.11 | 465 | 177.55 | 8.33 | 0.21, 0.27 |
| Zn2 | 280 (42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 341), 332 (5882) 341), 332 (5882) |
412 | 69.14 | 8.54 | 0.160 | 0.16, 0.07 | 480 | 147.14 | 9.13 | 0.25, 0.34 |
| L3 | 269 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 543), 334 (1195) 543), 334 (1195) |
402 | 82.40 | 8.62 | 0.028 | 0.20, 0.19 | 470 | 96.04 | 8.73 | 0.25, 0.29 |
| Zn3 | 281 (42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 808), 337 (2579) 808), 337 (2579) |
416 | 72.44 | 8.66 | 0.193 | 0.16, 0.07 | 495 | 104.34 | 9.64 | 0.23, 0.37 |
| L4 | 268 (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 246), 340 (1697) 246), 340 (1697) |
413 | 112.42 | 7.43 | 0.046 | 0.17, 0.13 | 500 | 155.62 | 7.55 | 0.30, 0.39 |
| Zn4 | 281 (45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 048), 350 (2829) 048), 350 (2829) |
436 | 105.03 | 8.08 | 0.362 | 0.17, 0.15 | 514 | 154.14 | 8.37 | 0.28, 0.38 |
| L5 | 270 (17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 357), 346 (2200) 357), 346 (2200) |
405 | 126.15 | 7.27 | 0.037 | 0.18, 0.13 | 480 | 152.64 | 8.91 | 0.29, 0.35 |
| Zn5 | 281 (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 371), 354 (19 371), 354 (19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 852) 852) |
422 | 65.15 | 8.04 | 0.278 | 0.16, 0.06 | 499 | 187.57 | 10.25 | 0.31, 0.38 |
In acetonitrile solution, the emission bands are observed at 393–413 nm, and 400–436 nm, respectively, for ligands L1–L5 (Fig. S9c and d†) and complexes Zn1–Zn5 (Fig. 7c and d). These bands exhibit deep blue luminescence emissions, and the Commission Internationale d'Eclairage (CIE) coordinates are summarized in Table 3. In acetonitrile solution, the maxima emission peaks are blue-shifted 65–87 nm compared with those in the solid state. These phenomena can be explained that the formation of the hydrogen bonds and π⋯π stacking interactions in the solid state, which can effectively decrease the HOMO–LUMO energy gap, and influence the ligand-centered π*–π transitions.44 Meanwhile, the full width at half-maximum (FWHM) of the emission band decreases from acetonitrile solution to the solid state (Table 3). All luminescence quantum yields of complexes Zn1–Zn5 in solution were measured by the optical dilute method of Demas and Crosby45 with a standard of quinine sulfate (Φr = 0.546) and are listed in Table 3. The luminescence quantum yield of Zn1–Zn5 in CH3CN is 0.095, 0.160, 0.193, 0.362 and 0.278, respectively, which is higher than that of the corresponding ligand. Particularly, the quantum yields of Zn4 (ΦF = 0.362) is 7.87-fold to L4 (ΦF = 0.046). This is because the ligands coordinated with zinc(II) ions increase the rigidity of the molecular edifice and reduce the loss of energy by radiationless thermal vibrations.46
The luminescence decay profiles of ligands L1–L5 and the corresponding zinc(II) complexes were measured at their optimal excitation wavelengths in the solid state and acetonitrile solution at 298 K (Fig. S10 and S11†). The detailed data are listed in Table S4.† A general trend is that the luminescence lifetimes for complexes Zn1–Zn5 either in the solid state or acetonitrile solution at 298 K are mostly longer than that of the corresponding ligands L1–L5. The most obvious observation is that the lifetime of Zn1 (τ = 9.15 μs) is 1.5-fold to the corresponding ligand L1 (τ = 6.10 μs) in acetonitrile solution. This is attributed to the more stable structure and interaction upon coordination.47 Meanwhile, the luminescence lifetimes of the ligand and corresponding complexes in the solid state (τ = 7.55–8.91 μs for L1–L5; τ = 8.37–10.25 μs for Zn1–Zn5) are longer than those in acetonitrile solution (τ = 6.10–8.62 μs for L1–L5; τ = 8.04–9.15 μs for Zn1–Zn5), which might be explained by the fact that there is a less polar nature in the solid-state environment.48
DFT calculations
The orbital distributions of the highest occupied molecular orbital (HOMO) and the lowest unoccupied molecular orbital (LUMO) energy of Zn1–Zn5 were calculated by DFT/B3LYP/6-31G (Fig. 8). Their optimized molecular structures are obtained from X-ray diffraction analyses of Zn1–Zn5. Therefore, we obtain information about their electron density distributions, energy gaps, and energy levels of HOMO and LUMO based on the calculated optimized molecular structures. | ||
| Fig. 8 Theoretically calculated spatial distributions and energies of the HOMO and LUMO levels of Zn1–Zn5. | ||
For all selected complexes, the HOMO electron density is mainly placed on the phenyl ring, while the LUMO electron density is mainly located on the pyridyl ring as shown in Fig. 8. Indeed, an analysis of the orbital distributions reveals a lesser participation of the Zn atom in the emission process for all complexes. These facts further demonstrate the π–π* nature of the intraligand charge transfer transitions in each case. The HOMO–LUMO gap is found to be 4.06 eV for Zn1 (H) and decreases to 3.99 to 3.94 to 3.70 to 3.80 eV for Zn2 (2-Me) to Zn3 (2,4-Me2) to Zn4 (2,4,6-Me3) to Zn5 (2-OMe), respectively. These results are consistent with the fact of electron-donating effect descended the energy difference between HOMO and LUMO.
Conclusion
In summary, five Zn(II) complexes with different alkyl substituent were synthesized and the effect of alkyl substitution on the structural as well as spectroscopic properties were also examined. It was found that complexes Zn1–Zn5 exhibit 3D → 1D supramolecular frameworks in pace with an increase in the number of alkyl substitution. The emission maximum wavelengths of complexes Zn1–Zn5 can be tuned in a large range of 400–514 nm due to the substitution with different alkyl substituent. This influence of ligand modification on the properties of the complexes was also confirmed by the theoretical calculations (DFT). The luminescent properties of zinc Schiff base complex are easily tunable by variation of the alkyl substitution. These kinds of zinc Schiff base complexes with supramolecular framework shows great promise for the preparation of a wide variety of transition-metal complexes with predictable and tunable properties.Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant 21371040 and 21571042), the National Key Basic Research Program of China (973 Program, No. 2013CB632900).References
- J. M. Lehn, Angew. Chem., Int. Ed. Engl., 1990, 29, 1304–1319 CrossRef.
- G. R. Desiraju, Crystal Engineering: The Design of Organic Solids, Elsevier, Amsterdam, 1989 Search PubMed.
- W. Y. Wong, L. Liu and J. X. Shi, Angew. Chem., Int. Ed., 2003, 42, 4064–4068 CrossRef CAS PubMed.
- W. Y. Wong, Coord. Chem. Rev., 2005, 249, 971–997 CrossRef CAS.
- L. L. Han, T. P. Hu, K. Mei, Z. M. Guo, C. Yin, Y. X. Wang, J. Zheng, X. P. Wang and D. Sun, Dalton Trans., 2015, 6052–6061 RSC.
- S. Yuan, Y. K. Deng and D. Sun, Chem.–Eur. J., 2014, 20, 10093–10098 CrossRef CAS PubMed.
- W. G. Lu, Z. W. Wei, Z. Y. Gu, T. F. Liu, J. Park, J. Park, J. Tian, M. W. Zhang, Q. Zhang, T. Gentle III, M. Bosch and H. C. Zhou, Chem. Soc. Rev., 2014, 43, 5561–5593 RSC.
- L. Fu, M. Pan, X. J. Kuang, C. Yan, K. Li, S. C. Wei and C. Y. Su, J. Mater. Chem. A, 2013, 1, 8575–8580 CAS.
- X. L. Hu, C. Qin, X. L. Wang, K. Z. Shao and Z. M. Su, Chem. Commun., 2015, 51, 17521–17524 RSC.
- Y. Q. Jiao, H. Y. Zang, X. L. Wang, E. L. Zhou, B. Q. Song, C. G. Wang, K. Z. Shao and Z. M. Su, Chem. Commun., 2015, 51, 11313–11316 RSC.
- Q. Y. Yang, M. Pan, S. C. Wei, K. Li, B. B. Du and C. Y. Su, Inorg. Chem., 2015, 54, 5707–5716 CrossRef CAS PubMed.
- O. M. Yaghi, H. Li, C. Davis, D. Richardson and T. L. Groy, Acc. Chem. Res., 1998, 31, 474–484 CrossRef CAS.
- J. R. Long and O. M. Yaghi, Chem. Soc. Rev., 2009, 38, 1213–1214 RSC.
- A. Y. Robin and K. M. Fromm, Coord. Chem. Rev., 2006, 250, 2127–2157 CrossRef CAS.
- A. Ali, G. Hundal and R. Gupta, Cryst. Growth Des., 2012, 12, 1308–1319 CAS.
- D. Natale and J. C. Mareque-Rivas, Chem. Commun., 2008, 425–437 RSC.
- M. Nishio, CrystEngComm, 2004, 6, 130–158 RSC.
- J. S. Field, O. Q. Munro and B. P. Waldron, Dalton Trans., 2012, 5486–5496 RSC.
- J. M. Lin, Y. X. Qiu, W. B. Chen, M. Yang, A. J. Zhou, W. Dong and C. E. Tian, CrystEngComm, 2012, 14, 2779–2786 RSC.
- R. F. Semeniuc, T. J. Reamer and M. D. Smith, New J. Chem., 2010, 34, 439–452 RSC.
- G. Mahmoudi, V. Stilinović, M. S. Gargari, A. Bauzá, G. Zaragoza, W. Kaminsky, V. Lynch, D. Choquesillo-Lazarte, K. Sivakumar, A. A. Khandari and A. Frontera, CrystEngComm, 2015, 17, 3493–3502 RSC.
- W. K. Lo, W. K. Wong, W. Y. Wong and J. P. Guo, Eur. J. Inorg. Chem., 2005, 3950–3954 CrossRef CAS.
- Q. H. Meng, P. Zhou, F. Song, Y. B. Wang, G. L. Liu and H. Li, CrystEngComm, 2013, 15, 2786–2790 RSC.
- G. M. Sheldrick, SHELXTL NT Crystal Structure Analysis Package, Version 5.10, Analytical X-ray System, Bruker AXS, Madison, WI, 1999 Search PubMed.
- H. Huang, Y. Wang, B. Wang, S. Zhuang, B. Pan, X. Yang, L. Wang and C. Yang, J. Mater. Chem. C, 2013, 1, 5899–5908 RSC.
- C. Han, G. Xie, J. Li, Z. Zhang, H. Xu, Z. Deng, Y. Zhao, P. Yan and S. Liu, Chemistry, 2011, 17, 8947–8956 CrossRef CAS PubMed.
- Y. Yuan, J. X. Chen, F. Lu, Q. X. Tong, Q. D. Yang, H. W. Mo, T. W. Ng, F. L. Wong, Z. Q. Guo, J. Ye, Z. Chen, X. H. Zhang and C. S. Lee, Chem. Mater., 2013, 25, 4957–4965 CrossRef CAS.
- P. Baran, R. Boca, M. Breza, H. Elias, H. Fuess, V. Jorik, R. Klement and I. Svoboda, Polyhedron, 2002, 21, 1561–1571 CrossRef CAS.
- X. Gong, Y. Y. Ge, M. Fang, Z. G. Gu, S. R. Zheng, W. S. Li, S. J. Hu, S. B. Li and Y. P. Cai, CrystEngComm, 2011, 13, 6911–6915 RSC.
- M. Shakir, M. Azam, Y. Azim, S. Parveen and A. U. Khan, Polyhedron, 2007, 26, 5513–5518 CrossRef CAS.
- A. W. Addison, T. N. Rao, J. Reedijk, J. van Rijn and G. C. Verschoor, J. Chem. Soc., Dalton Trans., 1984, 1349–1356 RSC.
- L. Yang, D. R. Powell and R. P. Houser, Dalton Trans., 2007, 955–964 RSC.
- Y. W. Dong, R. Q. Fan, P. Wang, L. G. Wei, X. M. Wang, S. Gao, H. J. Zhang, Y. L. Yang and Y. L Wang, Inorg. Chem., 2015, 54, 7742–7752 CrossRef CAS PubMed.
- M. Franks, A. Gadzhieva, L. Ghandhi, D. Murrell, A. J. Blake, E. S. Davies, W. Lewis, F. Moro, J. McMaster and M. Schröder, Inorg. Chem., 2013, 52, 660–670 CrossRef CAS PubMed.
- Y. W. Dong, R. Q. Fan, X. M. Wang, P. Wang, H. J. Zhang, L. G. Wei, W. Chen and Y. L. Yang, Cryst. Growth Des., 2016, 16, 3366–3378 CAS.
- F. Wu, H. Tong, Z. Li, W. Lei, L. Liu, W. Y. Wong, W. K. Wong and X. Zhu, Dalton Trans., 2014, 12463–12466 RSC.
- F. Wu, J. Li, H. Tong, Z. Li, C. Adachi, A. Langlois, P. D. Harvey, L. Liu, W. Y. Wong, W. K. Wong and X. Zhu, J. Mater. Chem. C, 2015, 3, 138–146 RSC.
- R. Y. Huang, H. Jiang, C. H. Zhu and H. Xu, RSC Adv., 2016, 6, 3341–3349 RSC.
- C. X. Ding, X. Rui, C. Wang and Y. S. Xie, CrystEngComm, 2014, 16, 1010–1019 RSC.
- X. Shi, G. S. Zhu, Q. R. Fang, G. Wu, G. Tian, R. W. Wang, D. L. Zhang, M. Xue and S. L. Qiu, Eur. J. Inorg. Chem., 2004, 185–191 CrossRef.
- C. Y. Xu, Q. Q. Guo, X. J. Wang, H. W. Hou and Y. T. Fan, Cryst. Growth Des., 2011, 11, 1869–1879 CAS.
- B. X. Dong and Q. Xu, Cryst. Growth Des., 2009, 9, 2776–2782 CAS.
- Y. H. Wu, L. Chen, J. Yu, S. L. Tong and Y. Yan, Dyes Pigm., 2013, 97, 423–428 CrossRef CAS.
- K. C. Stylianou, R. Heck, S. Y. Chong, J. Bacsa, J. T. A. Jones, Y. Z. Khimyak, D. Bradshaw and M. J. Rosseinsky, J. Am. Chem. Soc., 2010, 132, 4119–4130 CrossRef CAS PubMed.
- J. N. Demas and G. A. Crosby, J. Phys. Chem., 1971, 75, 991–1024 CrossRef.
- C. Maxim, F. Tuna, A. M. Madalan, N. Avarvari and M. Andruh, Cryst. Growth Des., 2012, 12, 1654–1665 CAS.
- H. Mandal, S. Chakrabartty and D. Ray, RSC Adv., 2014, 4, 65044–65055 RSC.
- K. D. Murugan and P. Natarajan, Eur. Polym. J., 2011, 47, 1664–1675 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 1477759, 1494305–1494308. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c6ra20377a |
| This journal is © The Royal Society of Chemistry 2016 |