Computational investigation of the interaction mechanism between the estrogen related receptor α and its agonists
Fuxing Lia,
Xianqiang Sunb,
Yingchun Caia,
Defang Fana,
Weihua Lia,
Yun Tang*a and
Guixia Liu*a
aShanghai Key Laboratory of New Drug Design, School of Pharmacy, East China University of Science and Technology, Shanghai 200237, China. E-mail: gxliu@ecust.edu.cn; ytang234@ecust.edu.cn; Fax: +86-21-64251033; Tel: +86-21-64250811
bDepartment of Biochemistry & Molecular Biophysics, Washington University School of Medicine, St. Louis, Missouri 63110, USA
First published on 27th September 2016
Abstract
Estrogen related receptor alpha (ERRα), the first identified orphan nuclear receptor, participates in the metabolism of body and adjusts some diseases such as breast cancer, osteoporosis and diabetes. Recent experiments have identified several ligands such as 7a, 7b, DK1 and DK3 toward ERRα as agonists, to have a therapeutic ability in reducing blood glucose, and impact the activity of ERRα receptor. Nevertheless, the detailed interaction modes between the agonists and the ERRα have not been fully understood, which will hinder the design and development of more potent agonists targeting ERRα for the treatment of diabetes. To address this issue, molecular docking and molecular dynamic simulation (MD) were performed to study how the four agonists regulate the behavior of ERRα. It was found that a proved residue Phe232 and some others such as Arg276, Phe286, His398 and Phe399 were the key residues in the ligand binding pocket. Hydrogen bonds between agonists and residues Arg276 and His398 and the pi–pi interactions between agonists and Phe232/Phe286/Phe399 both play important roles for agonist–ERRα binding. Interestingly, we observed that the agonists kept the binding of ERRα and its coactivator PGC-1α by stabilizing the site of helix12 (H12). All these findings are not only beneficial to understand the interaction mechanism between ERRα and its agonists, but also provide clues for designing novel and potent agonists of ERRα for the treatment of diabetes.
1 Introduction
Estrogen-related receptors (ERRs), members of nuclear receptor subfamily, contain three isoforms (ERRα, ERRβ and ERR γ).1,2 Though significant homology is shared between ERRs and estrogen receptors (ERs), ERRs don't bind with endogenous estrogen or any known natural ligands of ERs. Even more, recent functional genomic experiments have clearly demonstrated that ERRs and ERs display strict binding site specificities.3 ERRα is the first orphan nuclear receptor identified by a low stringency hybridization screen in 1988, which is expressed predominately in metabolically active tissues such as intestine, brown adipose tissue and skeletal muscles, and functions as a key regulator of energy metabolism.4Genetic and functional analysis have demonstrated that ERRα plays a critical role in regulating the energy of metabolism by controlling the transcriptional functions or protein expression of key components involved in tricarboxylic acid (TCA) metabolism,5 oxidative phosphorylation (OXPHOS) and glycolysis.6 Considering the simulation of energy and metabolism, ERRα can be studied as a potential therapeutic target in the treatment of such metabolic disorders as diabetes and obesity.7–10 Previous studies have shown that ERRα stimulates downstream biological function by recruiting coactivator such as peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α).11,12 And PGC-1α is an important coactivator in signal pathway of diabetes, so finding novel small molecules which can influence the activity of ERRα to solve metabolic diseases such as type 2 diabetes is intriguing.
Structural information of ERRα can provide us atomic details about the receptor and advance our understanding to the function of the receptor. Four crystal structures of ERRα have been resolved, which exhibit a canonical three layered “α-helical sandwich” fold composed of 12 α-helices (H1–H12) and two β-sheets.13 Two conformations of ERRα exist, the active one complexed with PGC-1α and the inactive one complexed with inverse agonists; both revealed a conserved structure in the binding site region. Nevertheless, significant divergences in both H12 and H3 were observed (Fig. 1). H12 is quite flexible, upon activation H12 binds with coactivator normally (the right figure of Fig. 1A), but in inverse agonist, a major conformational change takes place in the ligand binding domain (LBD) where H12 is repositioned to cap the ligand binding site, so the PGC-1α can't be combined for the position occupied by H12 (Fig. 1B and the left figure of Fig. 1A). The crystal structure14 of LBD of ERRα complexed with a coactivator peptide PGC-1α reveals a transcriptionally active conformation in the absence of a ligand. PGC-1α thereby plays a role in diverse processes including energy metabolism, mitochondrial biogenesis, and resistance to insulin in type 2 diabetes.15,16 Binding of activating ligand (agonist) to the ligand binding pocket can also trigger conformational changes and then result in coactivator recruitment. But the ligand binding pocket is highly occupied by residues Phe232, Phe286 and so on, which make it difficult to accommodate synthetic ligands without disrupting its constitutive active conformation (Fig. 2).
 | ||
| Fig. 1 (A) Two crystal structures of ERRα complex in PDB bank, namely, ERRα complexed with the inverse agonist XTC-790 (PDB entry code: 2PJL) and the active structure of ERRα complexed with PGC-1α (PDB entry code: 3D24). (B) States of H3 and H12 showed the most obvious differences between the inactive conformation (2PJL) and active conformation (3D24) of ERRα. | ||
 | ||
| Fig. 2 Chemical structures of ligands 7a, 7b, DK1 and DK3 and the ligand binding pocket of ERRα (PDB entry code: 3D24). The binding pocket is very small and locates in the deep site of ERRα protein. | ||
Several classes of inverse agonists have been identified by high-throughput screening and chemical synthesis.17–22 An X-ray crystallographic study demonstrated that the binding of inverse agonist XTC-790 with the ERRα LBD domain by interacting with Glu331, Val369 and one water in ERRα.23 All these demonstrated the inverse activation mechanism, but small molecular agonists of ERRα are lacking likely because of the tiny binding pocket of the ERRα LBD.14,24 Although the failure in identifying good ERRα agonists disappointed many scientists, several agonists, such as 7a, 7b, DK1, and DK3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25,26 have been found by the method of chemical synthesis finally and fortunately. All the four agonists (7a, 7b, DK1 and DK3) have a carbonyl group. 7a was an agonist that can effectively enhance ERRα-driven luciferase activity at 10.0 μM after a 24 h incubation, and 7b elevated the transcriptional function of ERRα by 2.7-fold at 10.0 μM because of the little difference between the two agonists. DK1 and DK3 were two agonists with similar scaffold to 7a and 7b. The EC50 values of DK1 and DK3 are not clear, but DK3 should be emphasized because of its specific selectivity to ERRα. The other three can also form complexes with ERRγ and ERs. Further studies showed these agonists could enhance the transcription of ERRα downstream target genes and improve the glucose and fatty acid uptake in C2C12 muscle cells, but it is not yet clear on the exact mechanism about how these agonists activate the receptor.
25,26 have been found by the method of chemical synthesis finally and fortunately. All the four agonists (7a, 7b, DK1 and DK3) have a carbonyl group. 7a was an agonist that can effectively enhance ERRα-driven luciferase activity at 10.0 μM after a 24 h incubation, and 7b elevated the transcriptional function of ERRα by 2.7-fold at 10.0 μM because of the little difference between the two agonists. DK1 and DK3 were two agonists with similar scaffold to 7a and 7b. The EC50 values of DK1 and DK3 are not clear, but DK3 should be emphasized because of its specific selectivity to ERRα. The other three can also form complexes with ERRγ and ERs. Further studies showed these agonists could enhance the transcription of ERRα downstream target genes and improve the glucose and fatty acid uptake in C2C12 muscle cells, but it is not yet clear on the exact mechanism about how these agonists activate the receptor.
In this study, in order to identify the activation mechanism, we concentrated on interactions between ERRα and four agonists (7a, 7b, DK1 and DK3). Molecular docking and molecular dynamics (MD) simulation were performed to investigate the binding interactions between the agonists and ERRα as well as the ligand-induced conformational changes of ERRα. With the initial conformation generated from docking, ten systems were constructed for subsequent MD simulations. In addition, we compared the differences between the specific selective agonist DK3 and the other three (7a, 7b and DK1). The results may avail us of understanding the activation mechanism of ERRα by agonists and provide clues for finding novel agonists of ERRα for the treatment of type 2 diabetes.
2 Materials and methods
2.1 Structure preparation
For ligands, the initial structures of the agonists 7a, 7b, DK1 and DK3 were geometrically optimized with the Macro Model module of the Schrödinger software package.27 The optimized conformer was thus obtained based on energy minimization, and was saved in PDB format for input into the docking protocol.For protein, the initial structure of ERRα was taken from Protein Data Bank (PDB). To date, there are four crystal structures available for ERRα. Two crystal structures complexed with inverse agonists were ignored, and one (PDB entry code: 3D24) of the two apo structures was chosen due to its high resolution (2.11 Å) finally.21 The crystal structure was then pretreated with the “Protein Preparation Wizard” workflow in Maestro version 9.3.28 In order to get complete structure, two missing loops were built using Prime.29
2.2 Molecular docking
We used the Autodock4.2 software package to predict the possible binding modes of agonists 7a, 7b, DK1 and DK3 to ERRα (3D24).30 The chemical structures of ligands and the binding pocket of ERRα were shown in Fig. 2. The center of the grid was placed on the ligand binding domain (LBD) and the residues within 20 Å around the center were defined as the binding pocket. Before starting the docking protocol, the target receptor and ligands were prepared using standard docking protocol and were saved as “PDBQT” format and Lamarckian Genetic Algorithm was used to generate 30 outputs for each docking run.31–33 In docking calculations, the target-ligand poses obtained were ranked using an energy based scoring function. After all outputs were clustered based on the root mean squared deviation (RMSD) values, the top pose conformation of docked ligand with the lowest energy in the biggest cluster was saved.2.3 MD simulation
| Simulation number | System name | Simulation time | Ligand | Coactivator |
|---|---|---|---|---|
| 1 | 7a_ERRα_PGC-1α | 200 ns | 7a | PGC-1α |
| 2 | 7a_ERRα | 200 ns | 7a | — |
| 3 | 7b_ERRα_PGC-1α | 200 ns | 7b | PGC-1α |
| 4 | 7b_ERRα | 200 ns | 7b | — |
| 5 | DK1_ERRα_PGC-1α | 200 ns | DK1 | PGC-1α |
| 6 | DK1_ERRα | 200 ns | DK1 | — |
| 7 | DK3_ERRα_PGC-1α | 200 ns | DK3 | PGC-1α |
| 8 | DK3_ERRα | 200 ns | DK3 | — |
| 9 | ERRα_PGC-1α | 200 ns | — | PGC-1α |
| 10 | ERRα | 200 ns | — | — |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-step energy minimization to relieve internal repulsions with the steepest descent algorithm with a maximum force of 10.0 kJ mol−1 nm−1 as the threshold. After energy minimization, NVT equilibration was conducted for 50 ps at 310 K. After stabilizing the temperature, NPT equilibration was performed for another 1 ns at 1 bar using the Parrinello–Rahman pressure coupling. The long-range electrostatics was set to the particle mesh Ewald algorithm,39 and constraints for all bonds with hydrogen atoms were applied using the LINCS algorithm. Following NVT and NPT equilibration, 200 ns production runs were carried out for ten systems using an NPT ensemble with a time step of 2 fs.
000-step energy minimization to relieve internal repulsions with the steepest descent algorithm with a maximum force of 10.0 kJ mol−1 nm−1 as the threshold. After energy minimization, NVT equilibration was conducted for 50 ps at 310 K. After stabilizing the temperature, NPT equilibration was performed for another 1 ns at 1 bar using the Parrinello–Rahman pressure coupling. The long-range electrostatics was set to the particle mesh Ewald algorithm,39 and constraints for all bonds with hydrogen atoms were applied using the LINCS algorithm. Following NVT and NPT equilibration, 200 ns production runs were carried out for ten systems using an NPT ensemble with a time step of 2 fs.2.4 MM-PBSA binding free energy calculation
The binding free energy of five PGC-1α/ERRα complexes was calculated using g_mmpbsa, which was developed to apply the MM-PBSA method to the GROMACS package. G_mmpbsa implemented the MM-PBSA method in the C programming language, removing any runtime dependency on external software such as DelPhi and APBS.44 In general terms, the binding free energy was conceptually summarized using the following equation:| ΔGbinding = Gcomplex − (Gprotein + Gligand) | (1) |
As described in previous publications,45 the free energy for each snapshot can be estimated from the following equation:
| ΔGbinding = ΔEMM + ΔGsolv − TΔS | (2) |
| ΔEMM = ΔEbonded + ΔEnonbonded | (3) |
| ΔEnonbonded = ΔEvdw + ΔEelec | (4) |
In eqn (4), ΔEvdw and ΔEelec represent van der Waals and electrostatic energy, respectively.
| ΔGsolv = ΔGpolar + ΔGnonpolar | (5) |
In current work, we adopted one of the most widely used nonpolar models, SASA-only nonpolar model (eqn (6)), for non-electrostatic solvation energy calculation.
| ΔGnonpolar = γ(SASA) + β | (6) |
3 Results and discussion
3.1 Docking analysis of agonists in human ERRα
In this study, molecular docking approach was used to determine the binding modes of the four agonists 7a, 7b, DK1 and DK3 in ERRα. Since no prior information is available about agonist–ERRα complexes in Protein Data Bank. Then we selected the conformations of the four agonists in estrogen related receptors with the most favorable energy score. The binding poses of the four ligands were shown in Fig. 3. The four small molecules share very similar scaffold, the main difference between 7a and 7b is the existence or absence of methyl or ethyl in the side chain. The difference between DK1 and DK3 lies in the chlorine atom on the side chain. However, obvious distinctions were observed for the binding poses between 7a and 7b, and between DK1 and DK3 (Fig. 3). The docked conformations of the ligands can be divided into two clusters. One cluster includes 7a and DK3; carbonyl oxygens of them located near to Phe232, while in the other cluster including 7b and DK1, the carbonyl oxygens located near to Phe286. From these two kinds of docking poses, we suspected that agonists with this scaffold have two low energy poses in protein pockets. In addition, benzene rings of agonists 7b and DK1 were surrounded by Phe232, Phe399 and Phe414 in the pocket domain. Two pi–pi interactions were found between the benzene rings of agonists 7b and DK1 and the side chains of residues Phe232 and Phe399. On the contrary, no pi–pi interaction was found in 7a and DK3 docking systems due to the long distance between the benzene ring on the ligand and the residues mentioned above. Based on the docking results, it can be concluded that the Phe232 and Phe399 of ERRα may play important roles for the binding of agonists.3.2 Stability of the systems and activation mechanism of the receptor
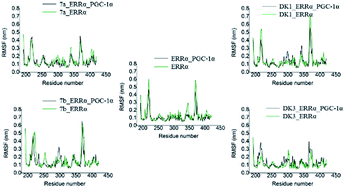
The RMSF values of the ten systems were also calculated which were shown in Fig. 4. The H2–H3 loop (loop between helix 2 and helix 3, residues 214–219) and H10–H11 loop (loop between helix 10 and helix 11, residues 367–373) in protein, which were broken in the crystal structure and repaired by the method of prime, had extremely larger fluctuation than any other parts including other loops. When agonists were complexed, the conformation changed slightly, for only some residues around the ligand binding pocket and H12 performed conformational changes. Ignoring the two unstable repaired loops, other minor fluctuations were caused by the movement of several key residues such as Phe232, Phe286, His398 and Phe399. Fluctuations of the ten systems are similar no matter the agonists and coactivator PGC-1α was complexed or not. But the fluctuation value in the two DK3 systems seems a little lower than the others, which implicates that systems complexed with DK3 are more stable.
From the results of interaction analysis (Fig. 5), we found that the 8 complex systems had different interaction modes. For the representational structure of system 7a_ERRα_PGC-1α, a hydrogen bond formed between the carbonyl oxygen of agonist 7a and the hydrogen atom of the imino group of the side chain of residue His398, and a pi–pi interaction was also found between the benzene ring of 7a and the residue Phe232. But for the structure of system 7a_ERRα, no hydrogen bond or pi–pi interaction were found. For agonist 7b, the snapshot of system 7b_ERRα_PGC-1α showed a hydrogen bond between the carbonyl oxygen of 7b and the hydrogen atom of the imino group of the side chain of residue His221, and a pi–pi interaction between the benzene ring of 7b and residue Phe232, too. But no hydrogen bond or pi–pi interaction was found in the snapshot of system 7b_ERRα. For agonist DK1, no hydrogen bond was found in systems DK1_ERRα_PGC-1α and DK1_ERRα, but two pi–pi interactions between DK1 and residues Phe286 and His398 were found in both systems with DK1. Finally, for agonist DK3, two hydrogen bonds formed between DK3 and ERRα. One hydrogen bond was between the carbonyl oxygen of DK3 and the hydrogen atom binding to the nitrogen atom of the guanidine group of Arg276, and the other formed between the nitrogen atom connecting with methyl in DK3 and the hydrogen atom binding to the other nitrogen atom of the guanidine group of Arg276. Besides the two hydrogen bonds, there was also a pi–pi interaction between the benzene ring of DK3 and Phe232 in system DK3_ERRα_PGC-1α. But no hydrogen bond was found in system DK3_ERRα, only the pi–pi interaction between DK3 and Phe286 existed. From above comparison, we can speculate that more hydrogen bonds and pi–pi interactions will form when PGC-1α is bound to ERRα simultaneously.
 | ||
| Fig. 5 Binding modes of the eight simulated systems complexed with agonists and key residues of the two simulated systems without agonists. | ||
The hydrogen bond interaction is crucial for the binding and specificity of ligands to the receptor, so hydrogen bond analysis along the whole simulation time and hydrogen bonding frequency analysis of the 20 ns equilibrium time were performed. During the whole simulation time, the agonist 7a formed only one hydrogen bond with the residue His398; 7b formed only one hydrogen bond with the residue His221. Differently, DK1 did not form any hydrogen bond with either of these two histidine residues. While DK3 formed three hydrogen bonds with other residues (Lys208, Ala211 and Arg276) of ERRα, but the hydrogen bond between DK3 and Ala211, with only 1% hydrogen bond occupancy, was extremely weak. Considering the results of hydrogen bonding frequency analysis in Table 2, and neglecting the systems complexed with DK1, which had no any hydrogen bond, we can further know that when PGC-1α is bound, the chance of forming hydrogen bond is larger no matter the agonist is 7a, 7b or DK3.
| Simulation number | System name | Residues related | Hydrogen bond occupancy |
|---|---|---|---|
| 1 | 7a_ERRα_PGC-1α | His398 | 40% |
| 2 | 7a_ERRα | His398 | 2% |
| 3 | 7b_ERRα_PGC-1α | His221 | 80% |
| 4 | 7b_ERRα | — | — |
| 5 | DK1_ERRα_PGC-1α | — | — |
| 6 | DK1_ERRα | — | — |
| 7 | DK3_ERRα_PGC-1α | Arg276 | 100% |
| Lys208 | 14% | ||
| Ala211 | 1% | ||
| 8 | DK3_ERRα | — | — |
Among the four agonists studied, DK3 is the only agonist which was known to selectively activate the ERRα. We further analyzed the behaviors of DK3 in the binding pocket of ERRα to discover the reason of the selectivity. Two obvious differences between DK3 and other three agonists appeared via above analyses. First, the lower RMSF value (Fig. 4) showed that the systems with DK3 are more stable. Second, we speculate that the hydrogen bond between DK3 and Arg276 (Table 2) contributes to the stability of DK3 simulated systems. What had already been proved was that the ligand would interact with Arg276 of ERRα, which had also been observed between ER/ERRγ and their selective ligands.46 It was clearly demonstrated that Arg276 played a real important role for the binding of agonists to ERRs.
From all these analyses, we can conclude that residues Phe232, which had been verified as a key residue via the mutation experiment,47 and Phe286 are the dominant residues which can form pi–pi interactions with ligands in most simulated systems, and His221, Arg276 and His398 are other three crucial residues to form hydrogen bonds with ligands in systems complexed with 7a, 7b and DK3, respectively. From this study, we knew those several residues around ligand binding pocket of ERRα are really important to impact the binding of ERRα and coactivator PGC-1α. All these interactions between key residues and ligands, especially the hydrogen bond of selective agonist DK3 can clearly show us the distinction of selective agonists of ERRα and help us find more novel ERRα agonists for the treatment of type 2 diabetes.
3.3 Impact of agonists on the binding of coactivator PGC-1α to ERRα
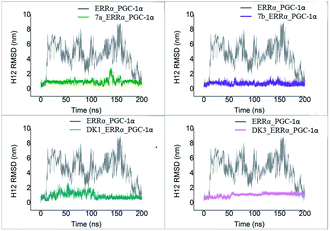
To test the stability of H12 in our simulations, RMSD of H12 for all the 200 ns simulation trajectory in each system was calculated and shown in Fig. 8. RMSD value of H12 in ERRα_PGC-1α simulated system without agonist reached a value of ∼5 to 8 Å, which is much higher than the other four ERRα_PGC-1α systems with agonists (about 1 Å), meaning H12 in the system of ERRα_PGC-1α without agonist was not stable at all, while H12 in the ERRα_PGC-1α systems with agonists were much more stable. Through comparing the RMSD of H12 in the five systems, we believed that agonists contribute to the stability of H12 in the estrogen related receptor, and then the stable conformation of H12 is helpful to the recruitment of the coactivator. These results support our inference that the agonists improve the binding of ERRα and the activation factor via stabilizing the level state of H12.
| System name | ΔGvdw | ΔGelec | ΔGsolv | ΔGSASA | ΔGbinding |
|---|---|---|---|---|---|
| ERRα_PGC-1α | −292.64 ± 0.79 | −793.89 ± 2.51 | 914.46 ± 3.36 | −34.33 ± 0.06 | −206.36 ± 2.43 |
| 7a_ERRα_PGC-1α | −209.40 ± 3.96 | −659.30 ± 19.06 | 668.66 ± 19.39 | −26.35 ± 0.35 | −225.83 ± 12.70 |
| 7b_ERRα_PGC-1α | −302.20 ± 1.97 | −816.30 ± 10.45 | 857.57 ± 13.51 | −36.79 ± 0.20 | −297.50 ± 6.93 |
| DK1_ERRα_PGC-1α | −275.38 ± 4.24 | −636.50 ± 12.57 | 686.31 ± 15.43 | −33.63 ± 0.41 | −259.35 ± 10.99 |
| DK3_ERRα_PGC-1α | −302.85 ± 2.21 | −660.89 ± 6.47 | 769.42 ± 9.26 | −36.25 ± 0.23 | −230.06 ± 8.32 |
From various analyses, we concluded that the small molecular agonists allosterically affect the stability of H12, then contribute to the recruitment of coactivator, which is similar to the verified activation mechanism in other agonists and nuclear receptor interaction systems.49–51
4 Conclusions
The newly-found four agonists of ERRα (7a, 7b, DK1 and DK3) were docked into the LBD of ERRα, which provided insights into the probable binding modes of agonists with ERRα. Subsequently, long-time MD simulations were performed for both apo-ERRα and ERRα/agonists complex systems, and binding mode of agonists to ERRα was depicted in detail. The docking results and interaction mode analysis of the MD trajectory displayed that Phe232, Arg276, Phe286, His398 and Phe399 around the binding pocket of ERRα are the key residues for the binding of agonists to ERRα. The results of MD simulations also reflected that the agonists may promote the binding of coactivator PGC-1α to ERRα by stabilizing the conformation and the site of H12. When agonists were absent, the site of H12 changed and hence hindered the binding of coactivator factor and protein, although the allosteric mechanism is not clear enough and more research is still needed, our model and findings may not only contribute to understand the binding mode of agonists to ERRα, but also supply useful information for searching more agonists of ERRα to help anti-diabetic drug design.Acknowledgements
We gratefully acknowledged the financial supports from the National Natural Science Foundation of China (Grant 81273438) and the Fundamental Research Funds for the Central Universities (Grant WY1113007).Notes and references
- S. Sanyal, J.-Y. Kim, H.-J. Kim, J. Takeda, Y.-K. Lee, D. D. Moore and H.-S. Choi, J. Biol. Chem., 2002, 277, 1739–1748 CrossRef CAS PubMed.
- S. N. Schreiber, D. Knutti, K. Brogli, T. Uhlmann and A. Kralli, J. Biol. Chem., 2003, 278, 9013–9018 CrossRef CAS PubMed.
- G. Deblois, J. A. Hall, M.-C. Perry, J. Laganière, M. Ghahremani, M. Park, M. Hallett and V. Giguère, Cancer Res., 2009, 69, 6149–6157 CrossRef CAS PubMed.
- D. J. Mangelsdorf, C. Thummel, M. Beato, P. Herrlich, G. Schütz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon and R. M. Evans, Cell, 1995, 83, 835–839 CrossRef CAS PubMed.
- A. Pedram, M. Razandi, B. Blumberg and E. R. Levin, FASEB J., 2016, 30, 230–240 CrossRef CAS PubMed.
- V. Giguère, Endocr. Rev., 2008, 29, 677–696 CrossRef PubMed.
- C. Handschin and V. K. Mootha, Drug Discovery Today, 2005, 2, 151–156 CAS.
- F. X. Soriano, M. Liesa, D. Bach, D. C. Chan, M. Palacín and A. Zorzano, Diabetes, 2006, 55, 1783–1791 CrossRef CAS PubMed.
- R. J. Patch, L. L. Searle, A. J. Kim, D. De, X. Zhu, H. B. Askari, J. C. O'Neill, M. C. Abad, D. Rentzeperis, J. Liu, M. Kemmerer, L. Lin, J. Kasturi, J. G. Geisler, J. M. Lenhard, M. R. Player and M. D. Gaul, J. Med. Chem., 2011, 54, 788–808 CrossRef CAS PubMed.
- V. Mootha, C. Handschin, D. Arlow, X. Xie, J. St Pierre, S. Sihag, W. Yang, D. Altshuler, P. Puigserver, N. Patterson, P. Willy, I. Schulman, R. Heyman, E. Lander and B. Spiegelman, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 6570–6575 CrossRef CAS PubMed.
- Y. Zhang, L. W. Castellani, C. J. Sinal, F. J. Gonzalez and P. A. Edwards, Genes Dev., 2004, 18, 157–169 CrossRef CAS PubMed.
- R. Bakshi, S. Mittal, Z. Liao and C. R. Scherzer, J. Parkinson's Dis., 2016, 5, 1–9 CrossRef PubMed.
- R. S. Savkur and T. P. Burris, J. Pept. Res., 2004, 63, 207–212 CrossRef CAS PubMed.
- J. Kallen, J.-M. Schlaeppi, F. Bitsch, I. Filipuzzi, A. Schilb, V. Riou, A. Graham, A. Strauss, M. Geiser and B. Fournier, J. Biol. Chem., 2004, 279, 49330–49337 CrossRef CAS PubMed.
- J. Lin, C. Handschin and B. M. Spiegelman, Cell Metab., 2005, 1, 361–370 CrossRef CAS PubMed.
- B. N. Finck and D. P. Kelly, J. Clin. Invest., 2006, 116, 615–622 CrossRef CAS PubMed.
- O. Lanvin, S. Bianco, N. Kersual, D. Chalbos and J.-M. Vanacker, J. Biol. Chem., 2007, 282, 28328–28334 CrossRef CAS PubMed.
- H. Greschik, J.-M. Wurtz, S. Sanglier, W. Bourguet, A. van Dorsselaer, D. Moras and J.-P. Renaud, Mol. Cell, 2002, 9, 303–313 CrossRef CAS PubMed.
- B. B. Busch, W. C. Stevens, R. Martin, P. Ordentlich, S. Zhou, D. W. Sapp, R. A. Horlick and R. Mohan, J. Med. Chem., 2004, 47, 5593–5596 CrossRef CAS PubMed.
- F. Wu, J. Wang, Y. Wang, T.-T. Kwok, S.-K. Kong and C. Wong, Chem.-Biol. Interact., 2009, 181, 236–242 CrossRef CAS PubMed.
- J. Wang, F. Fang, Z. Huang, Y. Wang and C. Wong, FEBS Lett., 2009, 583, 643–647 CrossRef CAS PubMed.
- S. Xu, X. Zhuang, X. Pan, Z. Zhang, L. Duan, Y. Liu, L. Zhang, X. Ren and K. Ding, J. Med. Chem., 2013, 56, 4631–4640 CrossRef CAS PubMed.
- J. Kallen, R. Lattmann, R. Beerli, A. Blechschmidt, M. J. J. Blommers, M. Geiser, J. Ottl, J.-M. Schlaeppi, A. Strauss and B. Fournier, J. Biol. Chem., 2007, 282, 23231–23239 CrossRef CAS PubMed.
- S. M. Hyatt, E. L. Lockamy, R. A. Stein, D. P. McDonnell, A. B. Miller, L. A. Orband-Miller, T. M. Willson and W. J. Zuercher, J. Med. Chem., 2007, 50, 6722–6724 CrossRef CAS PubMed.
- L. Peng, X. Gao, L. Duan, X. Ren, D. Wu and K. Ding, J. Med. Chem., 2011, 54, 7729–7733 CrossRef CAS PubMed.
- K. Ding, C. Wong, Z. Kang and X. Zhou, CN Pat., ZL200810026782.0, 2011.
- C. A. Romero, T. Grkovic, J. Han, L. Zhang, J. R. J. French, D. I. Kurtboke and R. J. Quinn, RSC Adv., 2015, 5, 104524–104534 RSC.
- S. Saxena, P. B. Devi, V. Soni, P. Yogeeswari and D. Sriram, J. Mol. Graphics Modell., 2014, 47, 37–43 CrossRef CAS PubMed.
- Y. Tsutsumi, Funkcialaj Ekvacioj, 1987, 30, 115–125 Search PubMed.
- X. Hou, J. Du, J. Zhang, L. Du, H. Fang and M. Li, J. Chem. Inf. Model., 2013, 53, 188–200 CrossRef CAS PubMed.
- G. M. Morris, D. S. Goodsell, R. S. Halliday, R. Huey, W. E. Hart, R. K. Belew and A. J. Olson, J. Comput. Chem., 1998, 19, 1639–1662 CrossRef CAS.
- M. F. Sanner, J. Mol. Graphics Modell., 1999, 17, 57–61 CAS.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
- C. R. Søndergaard, M. H. M. Olsson, M. Rostkowski and J. H. Jensen, J. Chem. Theory Comput., 2011, 7, 2284–2295 CrossRef PubMed.
- J. E. Del Bene, W. B. Person and K. Szczepaniak, J. Phys. Chem., 1995, 99, 10705–10707 CrossRef CAS.
- M. K. Gilson, K. A. Sharp and B. H. Honig, J. Comput. Chem., 1988, 9, 327–335 CrossRef CAS.
- X. Sun, H. Ågren and Y. Tu, J. Phys. Chem. B, 2014, 118, 10863–10873 CrossRef CAS PubMed.
- X. Sun, J. Cheng, X. Wang, Y. Tang, H. Ågren and Y. Tu, Sci. Rep., 2015, 5, 8066 CrossRef CAS PubMed.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- S. Wold, K. Esbensen and P. Geladi, Chemom. Intell. Lab. Syst., 1987, 2, 37–52 CrossRef CAS.
- M. A. Balsera, W. Wriggers, Y. Oono and K. Schulten, J. Phys. Chem., 1996, 100, 2567–2572 CrossRef CAS.
- M. L. Teodoro, G. N. Phillips and L. E. Kavraki, J. Comput. Biol., 2003, 10, 617–634 CrossRef CAS PubMed.
- F. Fratev, Phys. Chem. Chem. Phys., 2015, 17, 13403–13420 RSC.
- R. Kumari, R. Kumar and A. Lynn, J. Chem. Inf. Model., 2014, 54, 1951–1962 CrossRef CAS PubMed.
- L. Zeng, M. Guan, H. Jin, Z. Liu and L. Zhang, Chem. Biol. Drug Des., 2015, 86, 1438–1450 CAS.
- L. Wang, W. J. Zuercher, T. G. Consler, M. H. Lambert, A. B. Miller, L. A. Orband-Miller, D. D. McKee, T. M. Willson and R. T. Nolte, J. Biol. Chem., 2006, 281, 37773–37781 CrossRef CAS PubMed.
- W. Wei, A. G. Schwaid, X. Wang, X. Wang, S. Chen, Q. Chu, A. Saghatelian and Y. Wan, Cell Metab., 2016, 23, 479–491 CrossRef CAS PubMed.
- H. Greschik, M. Althage, R. Flaig, Y. Sato, V. Chavant, C. Peluso-Iltis, L. Choulier, P. Cronet, N. Rochel, R. Schüle, P.-E. Strömstedt and D. Moras, J. Biol. Chem., 2008, 283, 20220–20230 CrossRef CAS PubMed.
- C. Helsen and F. Claessens, Mol. Cell. Endocrinol., 2014, 382, 97–106 CrossRef CAS PubMed.
- L. Celik, J. D. D. Lund and B. Schiøtt, Biochemistry, 2007, 46, 1743–1758 CrossRef CAS PubMed.
- G. Costantino, A. Entrena-Guadix, A. Macchiarulo, A. Gioiello and R. Pellicciari, J. Med. Chem., 2005, 48, 3251–3259 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2016 |