Optimization of protein loaded PLGA nanoparticle manufacturing parameters following a quality-by-design approach
Abstract
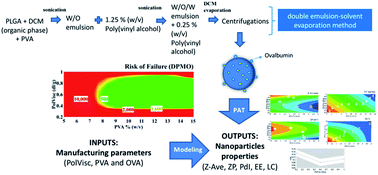
This paper discusses the development of a multivariate-based regression model for estimating the critical attributes to establish a design-space for poly(lactic-co-glycolic acid) (PLGA) nanoparticles formulated by a double emulsion–solvent evaporation method. A three-level, full factorial experimental design is used to assess the impact of three different manufacturing conditions (polymer viscosity, surfactant concentration and amount of model antigen ovalbumin) on five critical particle attributes (zeta potential, polydispersity index, hydrodynamic diameter, loading capacity and entrapment efficiency). The optimized formulation was achieved with a viscosity of 0.6 dl g−1, a surfactant concentration of 11% (w/v) in the internal phase and 2.5% (w/w) of ovalbumin. The design-space that is satisfied for nanoparticles with the targeted attributes was obtained with a polymer viscosity between 0.4 and 0.9 dl g−1, a surfactant concentration ranging from 8 to 15% (w/v) and 2.5% (w/w) of ovalbumin. The nanoparticles were spherical and homogenous and were extensively taken up by JAWS II murine immature dendritic cells without affecting the viability of these phagocytic cells. Better understanding was achieved by multivariate regression to control process manufacturing to optimize PLGA nanoparticle formulation. Utilization of multivariate regression with a defined control space is a good tool to meet product specifications, particularly over a narrow variation range.


 Please wait while we load your content...
Please wait while we load your content...