Multivariate curve resolution-assisted GC-MS analysis of the volatile chemical constituents in Iranian Citrus aurantium L. peel†
Abstract
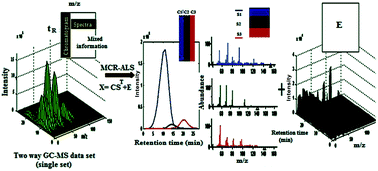
Multivariate curve resolution with alternating least squares optimization (MCR-ALS), as a soft modeling approach based on factor analysis, was proposed to recover the thorough gas chromatography-mass spectrometry (GC-MS) fingerprint analysis of volatile chemical constituents in Iranian Citrus aurantium L. peel. This technique for two-dimensional data was intended to resolve the overlapping and/or embedded GC-MS peaks into the pure chromatogram and mass spectrum of each chemical constituent, overcoming some challenging fundamental chromatographic problems occurring during GC-MS analysis of the Iranian C. aurantium peel chromatographic fingerprint, such as spectral background, baseline offset, and type of noise. In this way, the chromatographic fingerprints of Iranian C. aurantium peel are properly segmented to the appropriate chromatographic regions, and then MCR-ALS is used to achieve pure response profiles of the chemical constituents in each segment, as well as their relative concentrations. Retention indices, together with mass spectral profiles of pure chemical constituents, were considered for qualitative identification by matching against the standard ones through MS library searching; an overall volume integration (OVI) technique was also used for the semi-quantitative analysis (to obtain the relative concentrations of chemical constituents). GC-MS analysis of the C. aurantium L. peel, with the help of the proposed methodology, resulted in extending the number of identified constituents from 45 to 82 with concentrations higher than 0.01%. The lack of fit (LOF), percent of variance explained (R2) under the optimum conditions, and reverse match factor (RMF) were used for the assessment of the MCR-ALS solutions. The LOF values of MCR-ALS models were lower (12.0%) for all segment matrices with RMF values in the range 713–977 and R2 values higher than 97%. It was found that the major constituents of Iranian C. aurantium L. peel are limonene (72.89%), β-myrcene (9.06%), α-pinene (4.74%) and β-pinene (3.44%). It is concluded that the coupling hyphenated GC-MS measurements with the multivariate curve resolution-alternating least squares method is an effective and powerful strategy to solve current problems in GC-MS analysis, to obtain the required analytical selectivity in complex natural products.

 Please wait while we load your content...
Please wait while we load your content...