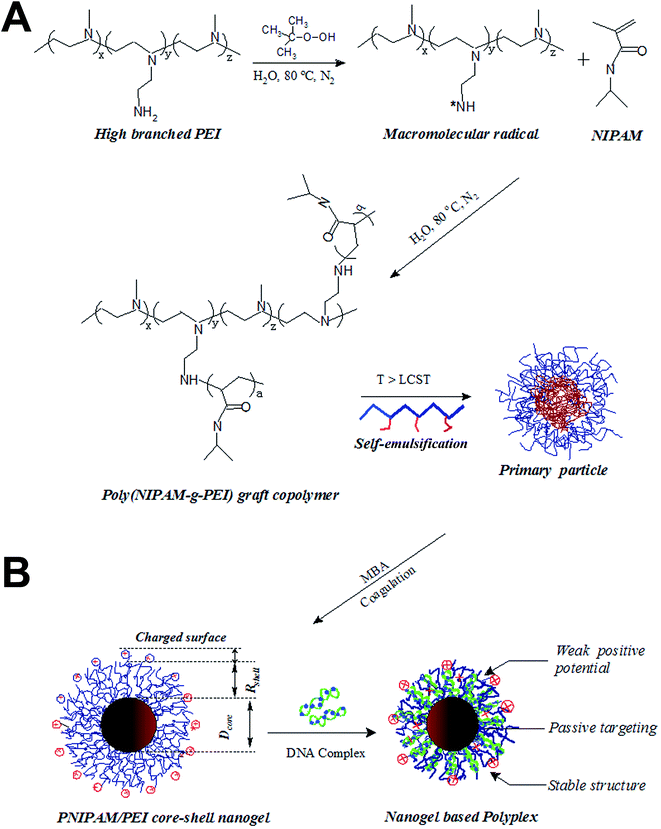
A nanogel with passive targeting function and adjustable polyplex surface properties for efficient anti-tumor gene therapy†
Haizhou Zhanga,
Qingbao Li*a,
Yingying Zhangb,
Yu Xiab,
Liang Yunc,
Qian Zhanga,
Tao Zhanga,
Xia Chenb,
Huaiwen Chenb and
Wei Lib
aDepartment of Cardiac Surgery, Shandong Provincial Hospital Affiliated to Shandong University, 324, Jingwu Road, Jinan 250021, China. E-mail: qingbaoli2004@sina.com; Tel: +86 0531 68773128
bInternational Joint Cancer Institute, The Second Military Medical University, 800 Xiangyin Road, Shanghai 200433, PR China
cDalian Institute for Drug Control, Yellow River Road 888A, Shahekou District, City of Dalian, Liaoning, China. E-mail: qingbaoli2004@sina.com; Tel: +86 0531 68773128
First published on 30th August 2016
Abstract
Non-cytotoxic vectors with high transfection efficiency and serum stability play a key role for successful gene delivery, which is strongly determined by polyplex structural properties and cellular affinity. In this study, through modifying the “gold-standard” transfection agent poly(ethylenimine) (PEI) with non-cytotoxic monomer N-isopropylacrylamide (NIPAM), we successfully developed a thermosensitive cationic PNIPAM/PEI nanogel. It has a crosslinked thermosensitive core and cationic shell with adjustable dimensions, surface potential and passive cellular targeting function. The polyplex properties such as size, surface potential, thermo-sensitivity and serum stability of the nanogel was systemically investigated by laser light scattering. It was found that the in vitro transfer efficiency by the nanogel was about two times higher than that of PEI, which was further enhanced about two time as T > VPTT due to the passive cellular targeting. The obviously enhanced in vitro cellular uptake, gene transfer efficiency and corresponding mechanism of the nanogel were then revealed by inverse fluorescent microscopy, a flow cytometer and confocal laser scanning microscopy. Its biodistribution and high intratumor accumulation were also evaluated in a Balb/c nude mice xenograft tumor model. Such a sophisticated nanogel significantly suppressed tumor growth with a volume 5 times smaller than that of PEI, which indicated its high potent for practical gene therapy.
1. Introduction
Since the first human gene therapy trials by Rosenberg in 1989, gene therapy has greatly improved the life quality of cancer patients as shown in clinical reports.1,2 Traditionally, guest genes were guided into the specific host cells by the physical transfer methods including electroporation, micro-injection or a gene gun, which are difficult to manipulate in practice.3 The virus vector with high transfer efficiency has been widely used in vitro and in vivo. However, its application is also forbidden due to the fatal accidents in clinical trials.4,5 Although the transfer efficiency of cationic liposomes was high in vitro, its in vivo application was also hindered by the multi-component, heterogeneous in size and morphology.6,7 The synthesized non-virus polymers are excellent alternatives for promoting gene transfection due to their low immunogenicity, tailorable structure and tunable size.8,9 Among cationic polymers, polyethylenimine (PEI, 25 kD) and its copolymers are “gold-standard” because of the “proton sponge” effects at intracellular pH (pH < 6.0).10 This protonated PEI promotes the negative DNA polyplex formation as well as the intracellular gene release. However, the high positive charged PEI can also lead to undesirable side cytotoxicity.11 In addition, on the cellular level, polyplex with weak positive potential and narrow size distribution is a key for successful endocytosis.12 So many efforts have been placed on the PEI chain modification.Strategies such as introducing non-cytotoxic or biodegradable monomers to PEI chains to form graft-/random-copolymers were employed in the literature to avoid the side effects.13–18 Most of the polyplexes formed with PEI copolymers was dominated by the positive/negative charged segments interaction (defined as N/P). It is thus difficult to optimize the polyplex surface potential and serum stability because the primary polyplex may aggregate resulting in low efficiency at low N/P. As N/P is increased, the repulsion among neighboring primary polyplexes increases as result of stable structure and high surface potential. But this large N/P ration will inevitably give rise to high side-cytotoxicity and low in vitro/vivo stability, which indicated that in vivo application of PEI based polyplexes could be limited by the unstable structure and surface properties.
Fortunately, the cationic nanogel, which is three-dimensional network with highly water-swollen structure, is a desirable non-virus vector for overcoming above mentioned obstacles. Its positive surface charge can be facilely tuned for gene condensing. While its cross-linked nanostructure ensures the in vitro/vivo polyplex stability.18–20 The PEG-c-PEI crosslinked network systems were developed for nucleoside analogs delivery. The system successfully reduced cytotoxicity of PEI and increased the nucleotides loading and release profile.21,22 The merit of this system could be further enhanced if it bearing some targeting function. The thermosensitive poly(N-isopropylacrylamide) (PNIPAM) was a well-known intelligent polymer, which hold the lower critical solution temperature (LCST ∼ 32 °C). Its LCST can be tuned by changing its chain backbone. Normally, the LCST will be increased when introducing hydrophilic monomer to the chain backbone. On the contrary, the LCST will be decreased by hydrophobically modification.14,15 The thermosensitive properties of nano carriers based on PNIPAM or its copolymers was well utilized to control the drug release in vitro/vivo. Previously, we and other groups have found that PNIPAM and its copolymers hold passive cellular targeting function as temperature was above its LCST.23–27 For creating a practical gene delivery system, it is still essential to disclose how to adjust the polyplex structural properties for promoting its in vitro/vivo transfer efficiency.12,28
In this study, the PNIPAM/PEI nanogel with crosslinked thermosensitive PNIPAM core and cationic PEI shell was finely fabricated which reduced cytotoxicity of PEI but enhancing the polyplex stability and cellular targeting. The detail polyplex properties including size (D), structure (Dcore/Rshell), surface charge (ξ), thermo-sensitivity (VPTT) and serum stability (PDI) was investigated by dynamic laser light scattering (DLS) and transmission electronic microscopy (TEM). The in vitro cellular uptake, transfer efficiency and corresponding mechanism were systematically investigated by inverse fluorescent microscopy (IFM), flow cytometer (FC) and confocal laser scanning microscopy (CLSM). In vivo biodistribution and intratumor accumulation were evaluated in Balb/c nude mice xenografted tumor by the IVIS® imaging system. The ricin A protein29 was used to evaluated the in vivo tumor inhibition.
2. Materials and methods
2.1. Materials
High branched poly(ethyleneimine) (PEI, Mw = 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1, 50 wt% in aqueous solution), N,N′-methylenebisacrylamide (MBA), N-isopropylacrylamide (NIPAM) were purchased from Aldrich. tert-Butyl hydroperoxide (TBHP, 70% in water) was obtained from Acros. 2,2′-Azobis-(2-amidinopropane) (V-50) was purchased from Wako Pure Chemical Industries. NIPAM was recrystallized three times from the mixture of toluene and hexane (1
000 g mol−1, 50 wt% in aqueous solution), N,N′-methylenebisacrylamide (MBA), N-isopropylacrylamide (NIPAM) were purchased from Aldrich. tert-Butyl hydroperoxide (TBHP, 70% in water) was obtained from Acros. 2,2′-Azobis-(2-amidinopropane) (V-50) was purchased from Wako Pure Chemical Industries. NIPAM was recrystallized three times from the mixture of toluene and hexane (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 v/v) mixture. MBA and V50 were recrystallized three times from the methanol. The hydrodated phosphotungstate (PTA), Dulbecco's phosphate buffered saline (PBS), fluorescein isothiocyanate (FITC), 4′,6-diamidino-2-phenylindole (DAPI), Cell Counting Kit-8 (CCK-8), heparin and bovine serum albumin (BSA, minimum 96%) (Sigma-Aldrich) were used as received. Quantum dot (QD, wavelength ∼605 nm, in aqueous solution) was purchased from Wuhan Jiayuan Quantun Dot Co. Ltd (China) and used as received. Water used in this study was purified by a water purification system, Milli-Q A10 (Millipore, Billerica, MA) at resistivity ∼18.2 MΩ cm.
5 v/v) mixture. MBA and V50 were recrystallized three times from the methanol. The hydrodated phosphotungstate (PTA), Dulbecco's phosphate buffered saline (PBS), fluorescein isothiocyanate (FITC), 4′,6-diamidino-2-phenylindole (DAPI), Cell Counting Kit-8 (CCK-8), heparin and bovine serum albumin (BSA, minimum 96%) (Sigma-Aldrich) were used as received. Quantum dot (QD, wavelength ∼605 nm, in aqueous solution) was purchased from Wuhan Jiayuan Quantun Dot Co. Ltd (China) and used as received. Water used in this study was purified by a water purification system, Milli-Q A10 (Millipore, Billerica, MA) at resistivity ∼18.2 MΩ cm.
2.2. Polymer synthesis and nanogel preparation
The graft copolymer PNIPAM-g-PEI was synthesized by the radical copolymerization in aqueous solution. The well-defined PNIPAM/PEI core–shell nanogel was in-suit formation during the polymerization process.30 Typically, the NIPAM monomer (0.48 g) was mixed with PEI (0.84 g) and MBA (0.5% weight percentage of the total monomer mass) in 25 mL Milli-Q water to form a transparent solution. The solution was bubbled with N2 for about 1 hour and then heated to 70 °C. TBHP (250 μL 0.01 M) was injected to the warm solution. Then the round bottom flask was sealed quickly under N2 bubbling. The solution was then heated up to 80 °C. The solution transferred from transparency to colloid about 15 min later. The reaction was kept at this temperature for about 6 hours under stirring. Then the reaction was stopped by open the system to air. The synthesis process was shown in Fig. 1. The cool white solution was purified by centrifugation three times. The upper solution was removed and the nanogel was re-dissolved in Milli-Q water. The concentration of this solution was calculated by lyophilization. A stock solution with C ∼ 10 mg mL−1 was stored at refrigerator for future use. The yield of the reaction was about 60% as evaluated by extraction method.18,312.3. Cell culture
Two cell lines, renal tubular epithelial cells (293T) was mainly used for cytotoxicity evaluation for PEI and PNIPAM/PEI nanogels. The human breast cancer cells (MDA-MB-231) was used for gene transfection evaluation and tumor xenograft. All cells were obtained from the American Type Culture Collection (ATCC). From the point of cell line, using the non-tumor breast cell (MCF10) is better than 293T. While as we known that the 293T cell holds the high gene transfection and protein reporting ability, which is widely used in evaluating the gene delivery efficiency by using the fluorescent protein model plasmid (pEGFP) as model gene.9,12,17 The cells were propagated and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS). Cultures were incubated in controlled atmosphere incubator at 37 °C with 5% CO2. DMEM and FBS were purchased from Gibco BRL (Grand Island, NY, USA). Before the in vitro transfection evaluation and other in vivo experiments, the cells were pre-cultured overnight until confluence was reached to 75%.2.4. Structure and properties of nanogel
| Cell viability% = [Asample − Ablank/(Acontrol − Ablank)] × 100 | (1) |
2.5. DNA condensation and polyplex characterization
Proportional amount of PEI and PNIPAM/PEI nanogel was dissolved in aqueous solution with different concentration. Then a small amount of above amplified pEGFP-N1 or pRA-EGFP DNA (amplification and purification was shown in S2, ESI†) (6 μg) was added to about 150 μL PEI and PNIPAM/PEI nanogel solution. The mixture was stirred for about 5 minutes and kept at room temperature for about 30 minutes as shown in Fig. 1B. The PEI-pRA-EGFP and PNIPAM/PEI nanogel polyplex was thus formed with different calculated N/P (∼3, 6, 7.5, 10, 15 and 30). DNase I enzyme degradation protection was conducted using the above mentioned PNIPAM/PEI nanogel polyplex solution. About 0.4 μL DNase I (10 unit per mL, in 2 μL reaction buffer (10×)) was added to 20 μL polyplex solutions (N/P ∼ 3, 6, 7.5, 10, 15 and 30). The polyplex was digested for about 5 minutes at 37 °C. About 2 μL 0.5 M ethylenediaminetetraacetic acid (EDTA) was added to each solution for stopping the enzyme digestion. After the inactivating, the temperature of the solution was heated to 80 °C and was kept for about 2 min. About 10 μL of each polyplex solution was sampled and analyzed by 0.7% agarose gel containing 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 GelRed Nucleic Acid Stain (Biotium, Inc). Gel electrophoresis was carried out in 1× TAE buffer (40 mM tris-acetate, 1 mM EDTA) at 100 V for 30 min in a electrophoresis apparatus (FR-250, Shanghai Furi). DNA bands were visualized by a UV lamp in a Gel image analysis system (FR-980, Shanghai Furi). The band 1 to 8 corresponded to DNA marker (Hindll band), the pDNA reference, the polyplex at 3, 6, 7.5, 10, 15, and 30, respectively. At the same time, about 12 μL 10 mg mL−1 heparin was added to another part of PNIPAM/PEI nanogel polyplex solution at different N/P and containing the DNAse1 enzyme and EDTA. The heparin has the highest negative charge which was used for the pDNA disassociation from the polyplex.31 This disassociation lasted for about 3 hours at room temperature. Then this polyplex solution was also analyzed by above mentioned 0.7% agarose gels which was used to evaluate the release profile. For further investigate the pDNA disassociation ability, the time dependence release profile was conducted by adding 10 mg mL−1 heparin to the polyplex solution. Then the accumulated release content was recorded at different time interval.
000 GelRed Nucleic Acid Stain (Biotium, Inc). Gel electrophoresis was carried out in 1× TAE buffer (40 mM tris-acetate, 1 mM EDTA) at 100 V for 30 min in a electrophoresis apparatus (FR-250, Shanghai Furi). DNA bands were visualized by a UV lamp in a Gel image analysis system (FR-980, Shanghai Furi). The band 1 to 8 corresponded to DNA marker (Hindll band), the pDNA reference, the polyplex at 3, 6, 7.5, 10, 15, and 30, respectively. At the same time, about 12 μL 10 mg mL−1 heparin was added to another part of PNIPAM/PEI nanogel polyplex solution at different N/P and containing the DNAse1 enzyme and EDTA. The heparin has the highest negative charge which was used for the pDNA disassociation from the polyplex.31 This disassociation lasted for about 3 hours at room temperature. Then this polyplex solution was also analyzed by above mentioned 0.7% agarose gels which was used to evaluate the release profile. For further investigate the pDNA disassociation ability, the time dependence release profile was conducted by adding 10 mg mL−1 heparin to the polyplex solution. Then the accumulated release content was recorded at different time interval.
PEI and PNIPAM/PEI nanogel based pRA-EGFP polyplex with N/P 3, 6 and 7.5 was prepared as above method. Then the polyplex was diluted by Milli-Q water with a concentration about 0.1 mg mL−1 for size characterization. The hydrodynamic diameter (D) and size distribution (PDI) of PEI and PNIPAM/PEI naongel based polyplex were determined at 25 °C by a dynamic laser light scattering (DLS) instrument of ZetaSizer (Nano-ZS) (Malvern Instruments, Worcestershire, UK). Each sample was measured in three runs and was calculated in average with cumulative errors smaller than 5%.32 At the same time, the zeta-potential in aqueous solutions was also measured by the Malvern Zetasizer 3000HS using 5 mM NaCl as the baseline.
The PNIPAM/PEI-pRA-EGFP polyplex solution was diluted by culture medium to diffident concentration for its cytotoxicity evaluation. For comparison, a parallel experiment, non-complexed pRA-EGFP plasmid was dissolved in culture medium, was also conducted. The non-tumor 293T cells were used for this cytotoxicity measurement. 293T cells were incubated in 96-well transparent plates (5000 cells per well in 0.1 mL medium) and incubated for overnight at 37 °C. Then the medium was removed. The cells were incubated with PNIPAM/PEI-pRA-EGFP polyplex or pRA-EGFP in DMEM medium with different concentration. It should be pointed out that the pRA-EGFP was used as the positive control. Moreover, for practical application of gene therapy, all the transferring and in vitro cytotoxicity evaluation was conducted in the medium with FBS. After about 24 hours later, the medium was removed and CC Kit-8 assay was used to measure the cell viability. The absorption of the samples in each well was measured by the microplate reader with wavelength 450 nm. The percentage of surviving cells was calculated according to above eqn (1). In this study, in order to check whether the nanogel system is no-cytotoxicity to the non-tumor cells, the incubation time was lasted as long as possible. On the contrary, in the investigation of the polyplex toxicity assay, 24 hours was enough.
2.6. In vitro transfection investigation
The in vitro transfection experiments were investigated by pEGFP-N1 DNA under different N/P (3, 6 and 7.5), different temperature (T < VPTT and T > VPTT) and different carriers (PEI and PNIPAM/PEI nanogel). All the transfection was conducted in the medium containing serum, which was close to the in vivo conditions. MDA-MB-231 cells were seeded onto 24-well plate at a density of 1.0 × 105 per well in 0.5 mL of DMEM containing 1% antibiotics and 10% FBS for 12 hours prior to transfection. PEI and PNIPAM/PEI nanogel based polyplex solutions containing different amount of pEGFP-N1 (with N/P ∼ 3, 6 and 7.5) was added to each well and were incubated for about 6 hours at 37 °C (T > VPTT). Then the medium was replaced with 0.5 mL of fresh DMEM supplemented with 10% FBS following with 30 hours culture resulting in a total transfection time about 36 hours. A parallel transfection experiment was conducted at culture temperature about 30 °C (T < VPTT) for about 6 hours, then transferred to 37 °C until the total culture time was 36 hours.On the other hand, the quantum dots (QDs in aqueous solution, from Wuhan Jiayuan Quantun Dot Co. Ltd, China) was used to evaluate the cellular internalization and intracellular pEGFP-N1 release. 10 μL QD solution was added to 200 μL PNIPAM/PEI nanogel solution (0.25 mg mL−1) and stirred for about 30 min. Then the unloaded QDs were removed by about 8 hours dialysis against water until the outside medium was clear. Noted here, the weight of nanogels for preparing such QD–nanogel complex was same as that for pDNA–nanogel polyplex. Moreover, both the QD–nanogel complex and the pDNA–nanogel polyplex (N/P ∼ 6) were incubated with same cell number (1.0 × 105 cells in 0.5 mL medium per well shown as following) at the same condition.
The inverse fluorescent microscopy and flow cytometer were used to record the fluorescent images and quantitatively analyze the fluorescent intensity. We use the positive ratio to evaluate the transfection. Because this parameter directly reflected the number of cells which contained the pEGFP-N1 or QD. MDA-MB-231 cells were seeded into 24-well microplates (1.0 × 105 cells per mL, 500 μL per well) and cultured for 12 hours. Then the cells were incubated with QD–nanogel complex and the pDNA–nanogel polyplex (N/P ∼ 6) in the medium with FBS 10%. After about 6 hours culture at 37 °C and 30 °C, the medium was replaced with 0.5 mL of fresh DMEM supplemented with 10% FBS following with 18 hours culture at 37 °C resulting in a total transfection time about 24 hours. Then the medium was removed and gently rinsed by PBS twice. Fluorescent microscopy for pEGFP-N1 (green) and QD (red) was recorded by the inverse fluorescent microscopy. After the cells were rinsed, digested and centrifuged, the positive ratio was collected using the FACScan flow cytometer (Becton Dickinson, San Jose, CA) at the same conditions.
2.7. Serum stability examination by DLS
The bovine serum albumin (BSA) was used to investigate the polyplex serum stable. The interaction between PNIPAM/PEI nanogel or PEI polyplex with high concentration BSA was examined by DLS instrument of ZetaSizer (Nano-ZS). The size and size distribution of PEI and PNIPAM/PEI nanogel based polyplex with a concentration ∼0.5 mg mL−1 and the BSA solution with a concentration ∼25 mg mL−1 in PBS (10×) was measured by DLS respectively. Secondly, above mentioned polyplex (C ∼ 0.5 mg mL−1 in water) was mixed with BSA (with C ∼ 25 mg mL−1 in PBS) under stirring for about 5 min. The size and size distribution of such mixture of polyplex and BSA was measured by the DLS. The serum stability was indicated by PDI.32 The properties such as size (D), distribution (PDI) and surface potential (ξ) was summarized in Table 1.| D/nm | PDI/a.u. | ξ/mV | VPTT/°C | |
|---|---|---|---|---|
| PEI | 450–5000 | >1 | 20 | — |
| PNIPMA/PEI | 300 | <0.5 | 0–5 | 32 |
2.8. Animal studies and tumor implantation
The experimental animals (Balb/c nude mice, female, 5 weeks, ∼20 g) were purchased from the Shanghai Experimental Animal Center of Chinese Academic of Sciences (Shanghai, P. R. China). Balb/c nude mice were maintained in a pathogen-free environment and allowed to acclimate for at least one week before tumor implantation. All of the experiments were performed in accordance with ethical guidelines under the protocols of the Committee on Animals of the Second Military Medical University (Shanghai China), which was approved by the Institutional Review Board of the Second Military Medical University.The above Balb/c nude mice were inoculated subcutaneously on the right shoulder with 5 × 106 MDA-MB-231 cells (in 100 μL culture medium) to develop xenografts tumor. After about 2 weeks, the volume of tumors reached about 50 mm3. The successful tumor xenograft model was demonstrated by tumor luminescent images. Briefly, mice were given an intraperitoneal injection of luciferin (Promega) at a dose of 150 mg kg−1. The tumor xenograft model was viewed by IVIS® Lumina II Imaging System (Xenogen), which was taken to capture the visible light photograph and luminescent image (S3, ESI†). As shown in S3 ESI,† immunofluorescence revealed a high EGFR expression in MDA-MB-231 tumor tissues, indicating that the tumor was successfully xenografted in vivo.27
2.9. In vivo distribution and tumor accumulation
In the study of biodistribution of polyplex, mice bearing MDA-MB-231 tumors were randomly assigned to 3 groups with 3 mice per group. The first group was used for background calibration which was injected PBS. The second group was used as reference by administration of PEI-FITC conjugates. The third group was used for study the administration site on the biodistribution of PNIPAM/PEI-FITC conjugates. Here, all the groups were administrated via tail vein (defined as i.v.). The total dosage of FITC was about 5 mg kg−1 and was administrated every day with three times. 24 hours later post final administration, the mice were anaesthetized by 1.5% isoflurane in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 O2/N2. The in vivo images were recorded under IVIS® imaging system (excitation 500 nm) and recorded by a built-in CCD camera. Mice were sacrificed after the photos taking. Then the organs heart, liver, kidney, tumor and spleen were excised which were also imaged by the same excitation wavelength. Finally, the organs were collected and immediately fixed in formalin for about 2 hours. The fixed organs were then frozen in tissue-Tek-OCT and cryosections. Frozen sections were cut at 10 μm sections and stored at −20 °C. After washing with PBS, sections were stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Sigma, Fluka Chemie, Buchs, Israel) and visualized by the CLSM (zeiss lsm 710, Germany).
2 O2/N2. The in vivo images were recorded under IVIS® imaging system (excitation 500 nm) and recorded by a built-in CCD camera. Mice were sacrificed after the photos taking. Then the organs heart, liver, kidney, tumor and spleen were excised which were also imaged by the same excitation wavelength. Finally, the organs were collected and immediately fixed in formalin for about 2 hours. The fixed organs were then frozen in tissue-Tek-OCT and cryosections. Frozen sections were cut at 10 μm sections and stored at −20 °C. After washing with PBS, sections were stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Sigma, Fluka Chemie, Buchs, Israel) and visualized by the CLSM (zeiss lsm 710, Germany).
2.10. Antitumor activity assay
Balb/c nude mice were inoculated subcutaneously with MDA-MB-231 cells (6 × 106 cells in 100 μL of PBS). Tumors were allowed to grow for about 2 weeks to reach volume around ∼50 mm3. Then the mice were randomly assigned to 3 groups with 5 mice per group. The first group was used as control by injection of 100 μL PBS. The second group was administrated the PEI based polyplex (loaded with pRA-EGFP). The third group was injected PNIPAM/PEI nanogel based polyplex (loaded with pRA-EGFP). The dosage for both PEI and nanogel group was about 200 μg per mice, which was administrated three times by every other day and 100 μL per injection. All the formulations were injected via tail vein. Tumor size was recorded every day by a digital vernier caliper. Both the serum stability and targeting issues were actually raised when the polyplex circulated in the blood. So in the experimental, administration of all drug formulations was via tail vein. The tumor volume (V) was calculated according to the following equation:25| Vtumor = LW2/2 | (2) |
2.11. Data analysis
Data were accumulated and expressed as the means ± standard deviation. Student's t-test was used to assess statistical significance of difference between group means, and p-values of 0.05 were considered statistically significant. ANOVA was used for comparing the difference among three or more groups in the experiments.3. Results and discussion
3.1. Core–shell structure tailoring and properties tuning
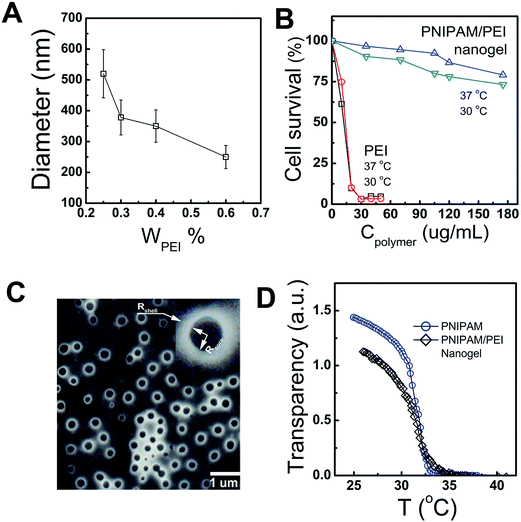
The PEI-g-PNIPAM copolymer was synthesized by radical graft copolymerization at 80 °C under N2 atmosphere as schemed in the process A in Fig. 1. The initiator tert-butyl hydroperoxide (TBHP) cleaved and gave raise the oxidant radicals at the reaction temperature. The –NH2 group along the high branched PEI (25 kD) was attacked by such oxidant resulted in the active radicals –NH*. Thus, the double bond on the NIPAM monomers was quickly imitated and produced a graft copolymer PEI-g-PNIPAM.31 All the impurities were removed from the particle dispersions through repeated centrifugation and decantation, followed by dialysis. It is well-known that PNIPAM grafted to the PEI chain backbone is thermosensitive polymer with a lower critical solution temperature (LCST) around 32 °C.33,34 The high reaction temperature (T ∼ 80 °C) leaded to an amphiphilic PEI-g-PNIPAM graft copolymer, which behavior likes macromolecular surfactant was schemed in Fig. 1A. The hydrophobic PNIPAM along the macromolecular surfactants (PEI-g-PNIPAM) aggregated each other to form primary emulsion particles.35 This process was called self emulsion as shown in Fig. 1B. Many neighboring primary particles further coagulated and crosslinked by the crosslinker (MBA) resulting in the PNIPAM/PEI nanogel with densely corsslinked PNIPMA core.9,17,35,36The size and surface potential of the PNIPMA/PEI nanogel play important role in gene condensing and transferring. How to regulate the morphology of nanogel is very important for its application. Principally, the number of the primer nanogel particle (Nparticle) in the emulsion polymerization are mainly determined by the macromolecular surfactant as shown in the following equation:35,36
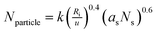 | (3) |
The thermosensitive properties of such PNIPAM/PEI nanogels was measured by the UV as shown in Fig. 2D. Both the PNIPAM homopolymer and the PNIPAM/PEI core–shell nanogel had the same volume phase transition temperature (VPTT ∼ 32 °C). The VPTT will be different from the PNIPAM homopolymer if some hydrophilic or hydrophobic monomers were introduced into PNIPAM chain backbone.37 Such similar VPTT value further indirectly reflected the well-defined core shell structure. As mentioned above, the temperature regulated VPTT can be used to enhance the cellular uptake indicating its passive cell targeting.14,15,21,24,38 On the other hand, a major problem associated with the golden standard PEI (25 kD) is its high toxicity, which is normally related to its surface potential.28 In this study, the cytotoxicity was successfully reduced by grafting with the bio-compatible NIPAMN. The potential was remarkably decreased from ∼40 mV to 20 mV (S4, ESI†). All these indicated that the well-defined PNIPAM/PEI nanogel with low positive charged PEI shell and thermosensitive PNIPAM core are suitable for anionic nucleic acids delivery. And the cellular uptaking could be enhanced as culture temperature was larger than its VPTT.28
3.2. Polyplex formation and properties
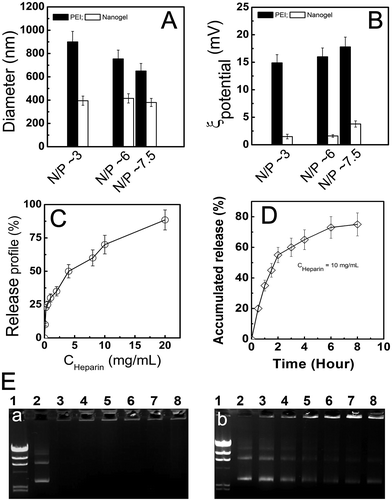
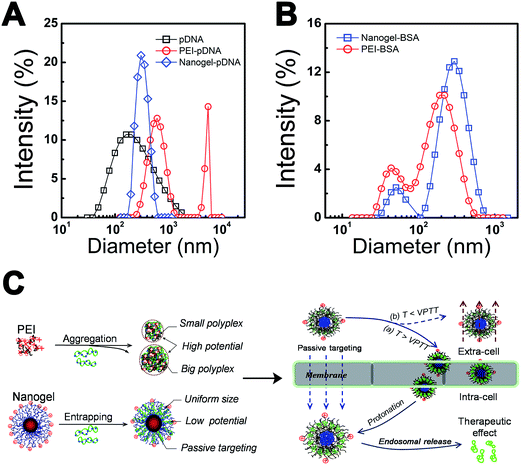
How to increase the transfection efficiency through regulating the complex between PNIPAM/PEI nanogel and pDNA is a key step because the in vitro/vivo transfection efficiency was strongly determined by the polyplex properties such as size and surface potential.11,28,39 Fig. 3A showed the N/P (3, 6 and 7.5) dependence of the PEI and PNIPAM/PEI pRA-EGFP polyplex size (D). At same N/P, the size of PEI based polyplex was about 2 times larger than that of PNIPAM/PEI nanogel. The D of PEI based polyplex decreased a little with N/P increase, which was attributed to the stronger condensation effect at high charged PEI content. On the contrary, the size of nanogel based polyplex had little changes, which indicated that its high stability. In addition, the surface potential of PEI based polyplex was also 10 times higher than that of nanogel based polyplex (Fig. 3B). The higher potential will lead to unstable structure and low transfection for the serum proteins absorption. Additionally, this highly charged polyplex surface is high cytotoxic to cells.11Fig. 3C showed the pDNA release profile from the nanogel at N/P ∼ 7.5. The anionic heparin was used to disassociate the pDNA by its negative charged chain. The pDNA release content increased as the heparin concentration increase indicated that the release profile was tunable. The well-defined core–shell PNIPAM/PEI nanogel still holds the desirable gene transfection efficiency due to the large amount of positive charge on the PEI chain surrounding the PNIPAM core. Fig. 3D showed the time dependence of pDNA release profile at a given heparin concentration (∼10 mg mL−1). It was found that the pDNA was released out from the nanogel by a steady and sustained mode. Both the heparin concentration and time dependence of the pDNA release profile clearly indicate the nanogel was an excellent candidate for gene delivery.18,31 Fig. 3E showed the enzyme degradation protection by PNIPAM/PEI nanogel at N/P ∼ 3, 6, 7.5, 10, 15 and 30, which corresponded to bands 3 to 8 (a). Noted here, the band 1 was marker, while band 2 was naked pDNA. No degradation was found from bands 3 to 8, which indicated the pDNA entrapped in the nanogel by large amount of positive PEI chain in the shell. These arms embedded pDNA and protected from the DNase I digestion. On the contrary, the naked pDNA was almost degraded resulted in a long white tail (band 2). However, when the heparin (∼5 mg mL−1) was added to the PNIPAM/PEI nanogel based polyplex solution. The pDNA was released out again as shown in b. This was attributed to the pDNA condensed and collapsed in the PEI shell was counteracted by the strong ionic heparins.18,31 The successful transferring and releasing was further evaluated by the cytotoxicity against to tumor cells (S5, ESI†). This heparin induced pDNA release illustrated that the thermosensitive core–shell nanogel held high pDNA loading and enzyme protection capacity.
3.3. In vitro transfection and intracellular release evaluation
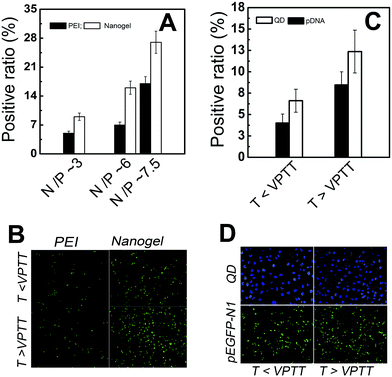
It should be pointed out that all the transfer experiments were conducted in serum containing medium because many serum and proteins appeared in the in vivo gene delivery.9,12,28 Fig. 4A showed the N/P dependence of in vitro transfection efficiency by MDA-MB-231 cells (evaluated by pEGFP-N1 DNA) for PEI and PNIPAM/PEI nanogel. As the N/P increased from 3 to 7.5, the transfection efficiency of PEI increased from about 6 to 17. However, for the nanogel, the transfection increased from about 10 to 30. At a given N/P, the transfection efficiency by nanogel was 2 times higher than that of the PEI. The reason for the lower transfection efficiency of PEI was mainly attributed to the high surface potential of naked PEI based polyplex, which resulted in aggregation with serum proteins with large polyplex size (Fig. 3A).9,12 In addition, at the given temperature (T > VPTT or T < VPTT), the transfection efficiency of PEI was lower than that of PNIPAM/EPI nanogel. But for PEI based polyplex, temperature had little effect on the transfection. But the transfection efficiency by nanogel at T > VPTT was much higher than that as T < VPTT (Fig. 4B).25–27 This was attributed to the temperature enhanced cell uptaking. Such temperature regulated passive targeting phenomenon was also reported by others.14,15,21,22,40As the pDNA was transported into the tumor cells. The intracellular gene release and reporting strongly affected the therapeutic index.11,41 Fig. 4C showed the temperature dependence of positive ratio of QD and pDNA uptaking by MDA-MB-231 cells. At the same transfer conditions and given temperature (T > VPTT or T < VPTT), the positive ratio of QD was about 60% higher than that of pDNA (N/P ∼ 6). While both the positive ratio of QD and pDNA transferred at T > VPTT was about 2 times higher than that at T < VPTT. At same transfer condition, the number of polyplex (contained either QD or pDNA) entered into the cells should be similar. The positive ratio of QD directly reflected the number ratio of cells uptaking nanogels to total cells. However, Fig. 4C showed that the positive ratio of the pEGFP-N1 was smaller than that of QD. This indicated that the pEGFP-N1 might not be completely released out. It might also due to low intracellular gene reporting.11 The effects of temperature and intracellular releasing on the transfection was directly visualized by the fluorescent image as shown in Fig. 4D. At the given N/P ∼ 6, it was found that efficiency at 36 hours (Fig. 4A) was higher than that at 24 hours (Fig. 4C, T < VPTT). This evidence indicated that the intracellular pDNA releasing and/or reporting strongly affected the transfer efficiency.9,12,28 The hydrophobic transition of thermosensitive PNIPAM core can enhance the gene transfection due to the passive targeting function as illustrated above.25–27
3.4. In vivo biodistribution and tumor accumulation
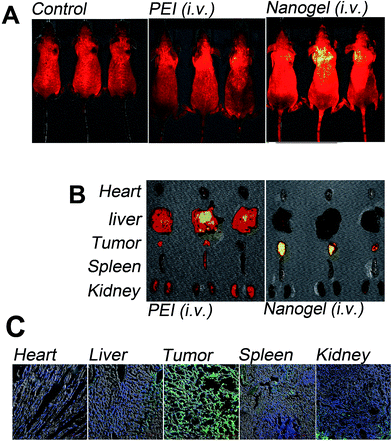
The in vivo polyplex distribution and tumor accumulation was evaluated in nude mice bearing breast cancer (MDA-MB-231 cells). The successfully established solid-tumor xenograft model was confirmed by the IVIS® imaging system shown in S3, ESI.†9,25 Then PBS, FITC-labeled PEI and -nanogel (PEI-FITC, PNIPAM/PEI-FITC) was intravenously injected into the MDA-MB-231 human breast tumor-bearing mice. About 24 hours post the final administration, the distribution and tumor accumulation was visualized by the IVIS® imaging system as shown in Fig. 5A.28 From the living animal fluorescent images, it was found that a little FITC accumulated in the solid tumor as delivered by PEI. This low tumor accumulation of PEI based polyplex was mainly due to its low serum stability.9,12 This large uniform size and high cytotoxic polyplex mainly accumulated in liver as shown in Fig. 5B. Such high liver accumulation was reasonable because liver was the main organ for detoxification.42On the contrary, the tumor accumulation of PNIPAM/PEI-FITC was obviously higher than that of PEI. Such enhanced tumor accumulation was also confirmed by the organs' fluorescent images shown in Fig. 5B. At the same conditions, its high intratumor accumulation was mainly due to the high serum stability. This serum stability prolonged the PNIPAM/PEI nanogel based conjugates in vivo circulation which resulted in enhanced permeability and retention (EPR) effects.9,27,43 From Fig. 5B, it was easy to find that the tumor accumulation of PNIPMA/PEI-FITC was obviously higher than that of PEI-FITC, which was further confirmed by the frozen section as shown in Fig. 5C. The cell nucleus was stained by DAPI as shown in blue. The green color in the section generated from the FITC molecules. The high serum stability, narrow polyplex size distribution and crosslinked structure enhanced the high tumor accumulation of PNIPAM/PEI nanogel.
3.5. In vivo anti-tumor activity evaluation
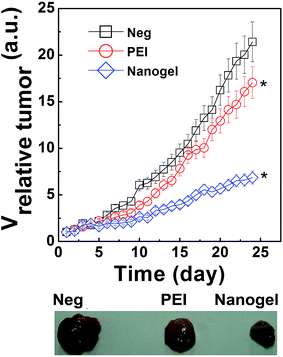
The Ricin A encoding plasmid DNA (pRA-EGFP) constructed in our lab was complexed with PNIPAM/PEI nanogel and PEI at the same N/P.29 As the tumor grown to about 50 mm3, the polyplex was i.v. administrated with a total dosage about 200 μg per mice. The relative tumor volume was recorded every day as shown in Fig. 6. The control group (namely, the Neg. group injected with PBS) was used as reference to evaluate the therapeutic index of polyplex. The relative tumor volume of this Neg. group was about 23, which was a little larger than that of PEI group (∼16). On the contrary, the tumor volume of group treated by PNIPMA/PEI nanogel based polyplex was much smaller than that of the Neg. and PEI groups. The tumor volume of PEI group was about 3.5 times larger than that of nanogel group which clearly indicated that the PNIPAM/PEI nanogel based polyplex successfully accumulated into the tumor by the EPR effect.9,28 The DNA was successfully transferred into the tumor cells by the nanogel due to the its well regulated polyplex properties such as the uniform size stable structure.12 Here, the intracellular pDNA release intratumor was attributed to the “proton sponge” hypothesis of PEI shell. The amine groups inside the shell are not fully protonated at physiological pH.18,19 As the polyplex was accumulated in tumor, internalized by the tumor cells and entrapped in the endo/lysosomal compartment with low pH. The protonation resulted in an expansion of PEI chain due to intra-chain charge repulsion.44 This causes an osmotic swelling and breaking of the polyplex with pRA-EGFP DNA releasing into the cytoplasm to exert its therapeutic effect with VNeg > VPEI. > Vnanogel. Taken together, it is easy to conclude that the thermosensitive PNIPAM/PEI nanogel showed high potent in the practical gene therapy.3.6. Effects of polyplex properties on the gene transferring and tumor growth inhibition
It is helpful to find that the effects of polyplex properties such as carrier's structure, composition, size, size distribution and surface potential strongly affect the cellular level gene transfection, in vivo distribution and tumor accumulation. How to promote the polyplex in vitro/vivo performance by regulating these properties is an important issue for gene therapy.11,28,45 Fig. 7A showed the size and size distribution of PEI and PNIPAM/PEI nanogel based polyplex. Two peaks (∼400 nm and 2000 nm) appeared in PEI-pDNA polyplex solution. The size of EPI was smaller than 10 nm. The large size and broad distribution (PDI) of PEI-pDNA polyplex indicated some inter-polyplex aggregation during the particle formation process.25,46 On the contrary, for the PNIPAM/PEI-pDNA polyplex, there was just one narrow peak (∼300 nm), which was similar to the size of nanogel. Both the small size and narrow PDI indicated that no inter-polyplex aggregation in the solution.9,28For testing the polyplex stability, the polyplex was mixed with serum proteins (BSA) at high concentration (∼25 mg mL−1) and indicated by dynamic laser light scattering (DLS). A very large size with large PDI was found in PEI based polyplex solution shown in Fig. 7B. The based polyplex formation with PEI was dominated by neutralization and aggregation, which leaded to high charge surface (∼20 mV) with strong interaction with BSA due to highly positive charged surface.12,32,45 The inter-polyplex aggregation and BSA binding resulted in large polyplex size distribution and low serum stability which induced low gene transfection and in vivo tumor accumulation as shown in Fig. 7C.9,12 On the contrary, for the nanogel based polyplex, the pDNA was absorbed and subsequently entrapped into nanogel resulted in narrow size, low surface potential and stable structure. So the peaks for BSA (∼10 nm) and nanogel based polyplex (∼300 nm) were well separated indicating no BSA absorption.27,46,47 The related low surface charge and inner crosslinked core contributed to nanogel's narrow PDI and high serum stability.12,17,20 Moreover, introducing thermosensitive PNIPAM further leaded to the passive cellular targeting ability Fig. 7C. Thus, the cellular uptake of nanogel based polyplex was obviously enhanced via the passive targeting function at T > VPTT. On the contrary, the cellular uptake will decrease at T < VPTT which was schemed in the right panel in Fig. 7C. In the intracellular level, the pDNA was released out by the protonation of the PEI chain at low pH.10 The therapeutic effect was consequently enhanced by the passive targeting and successfully intracellular pH regulated DNA release. Thus, by finely chemical synthesis and structural tailoring, the polyplex properties such as size (D), structure (Dcore/Rshell), surface charge (ξ), thermo-sensitivity (VPTT) and serum stability (PDI) (Table 1) was facilely regulated which resulted in low side cytotoxicity, high gene transfection and tumor growth inhibition.
4. Conclusion
Using the conventional radical graft copolymerization, we synthesized an amphiphilic graft copolymer PEI-g-PNIAPM at the temperature 80 °C. As in-suit crosslinked by MBA, a well-defined non-cytotoxicity core–shell PNIPAM/PEI nanogel was prepared in the emulsion polymerization. The DLS and TEM clearly showed its well-defined core–shell structure, the narrow size (D) and size distribution (PDI). In addition, the plasmid pRA-EGFP with green fluorescent protein GFP and toxin Ricin A gene were constructed and used to evaluate the in vivo transfer efficiency and tumor growth inhibition. The nanogel successfully promoted the enzyme protection with tunable intracellular pDNA release. The in vitro transfection by such core–shell nanogel was much higher than that of PEI. This efficiency was further promoted as T > VPTT due to T induced cellular targeting. In addition, comparing with PEI, the nanogel also showed much higher in vivo tumor accumulation. The tumor inhibition by pRA-EGFP delivered by PNIPAM/PEI polyplex was obviously promoted due to the fine size (D), structure (Dcore/Rshell), surface charge (ξ), thermo-sensitivity (VPTT) and serum stability (PDI) tailoring. Based on this systemically in vitro/vivo study and detail mechanism illustration, the thermosensitive PNIPAM/PEI nanogel can be expected as the efficient platform for practical gene delivery.Acknowledgements
This work was financially supported by Projects of Medical and Health Technology Development Program in Shandong province 2014WS0094, Ministry of Science and Technology of China (2012CB934002), the National Natural Science Foundation of China including the project (81302363, 31470964). In this work, Li Q. and Li W. contributed equally.References
- S. A. Rosenberg, P. Aebersold, K. Cornetta, A. Kasid, R. A. Morgan, R. Moen, E. M. Karson, M. T. Lotze, J. C. Yang and S. L. Topalian, N. Engl. J. Med., 1990, 323, 570–578 CrossRef CAS PubMed.
- M. L. Edelstein, M. R. Abedi and J. Wixon, J. Gene Med., 2007, 9, 833–842 CrossRef PubMed.
- J. M. E. scoffre, T. Portet, C. Favard, J. Teissié, D. S. Dean and M. P. Rols, Biochim. Biophys. Acta, 2011, 1808, 1538–1543 CrossRef PubMed.
- E. Check, Nature, 2002, 420, 116–118 CrossRef CAS PubMed.
- S. E. Raper, N. Chirmule, F. S. Lee, N. A. Wivel, A. Bagg, G. P. Gao, J. M. Wilson and M. L. Batshaw, Mol. Genet. Metab., 2003, 80, 148–158 CrossRef CAS PubMed.
- X. Guo and L. Huang, Acc. Chem. Res., 2012, 45, 971–979 CrossRef CAS PubMed.
- K. Un, S. Kawakami, R. Suzuki, K. Maruyama, F. Yamashita and M. Hashida, Biomaterials, 2010, 31, 7813–7826 CrossRef CAS PubMed.
- A. Tamura and N. Yui, Biomaterials, 2013, 34, 2480–2491 CrossRef CAS PubMed.
- W. Li, H. Li, J. Li, H. Wang, H. Zhao, L. Zhang, Y. Xia, Z. Ye, J. Gao, J. Dai, H. Wang and Y. Guo, Int. J. Nanomed., 2012, 7, 4661–4677 CrossRef CAS PubMed.
- M. Neu, D. Fischer and T. Kissel, J. Gene Med., 2005, 7, 992–1009 CrossRef CAS PubMed.
- S. M. Moghimi, P. Symonds, J. C. Murray, A. C. Hunter, G. Debska and A. Szewczyk, Mol. Ther., 2005, 11, 990–995 CrossRef CAS PubMed.
- D. W. Pack, A. S. Hoffman, S. Pun and P. S. Stayton, Nat. Rev. Drug Discovery, 2005, 4, 581–593 CrossRef CAS PubMed.
- X. Bao, W. Wang, C. Wang, Y. Wang, J. Zhou, Y. Ding, X. Wang and Y. Jin, Biomaterials, 2014, 35, 8450–8466 CrossRef CAS PubMed.
- M. Türk, S. Dinçer and I. G. Yuluğ, J. Controlled Release, 2004, 28, 325–340 CrossRef PubMed.
- H. S. Bisht, D. S. Manickam, Y. You and D. Oupicky, Biomacromolecules, 2006, 7, 1169–1178 CrossRef CAS PubMed.
- P. Lemieux, S. V. Vinogradov, C. L. Gebhart, N. Guérin, G. Paradis, H. K. Nguyen, B. Ochietti, Y. G. Suzdaltseva, E. V. Bartakova, T. K. Bronich, Y. St-Pierre, V. Y. Alakhov and A. V. Kabanov, J. Drug Targeting, 2000, 8, 91–105 CrossRef CAS PubMed.
- Y. Yue, F. Jin, R. Deng, J. Cai, Y. Chen, M. C. Lin, H. F. Kung and C. Wu, J. Controlled Release, 2011, 155, 67–76 CrossRef CAS PubMed.
- H. Mimi, K. M. Ho, Y. S. Siu, A. Wu and P. Li, J. Controlled Release, 2012, 158, 123–130 CrossRef CAS PubMed.
- S. E. Averick, E. Paredes, A. Irastorza, A. R. Shrivats, A. Srinivasan, D. J. Siegwart, A. J. Magenau, H. Y. Cho, E. Hsu, A. A. Averick, J. Kim, S. Liu, J. O. Hollinger, S. R. Das and K. Matyjaszewski, Biomacromolecules, 2012, 13, 3445–3449 CrossRef CAS PubMed.
- W. Li, Q. Guo, H. Zhao, L. Zhang, J. Li, J. Gao, W. Qian, B. Li, H. Chen, H. Wang, J. Dai and Y. Guo, Nanomedicine, 2012, 7, 383–392 CrossRef CAS PubMed.
- S. Vinogradov, E. Batrakova and A. V. Kabanov, Colloids Surf., B, 1999, 16, 291–304 CrossRef CAS.
- S. V. Vinogradov, A. D. Zeman, E. V. Batrakova and A. V. Kabanov, J. Controlled Release, 2005, 107, 143–157 CrossRef CAS PubMed.
- A. Schwerdt, A. Zintchenko, M. Concia, N. Roesen, K. Fisher, L. H. Lindner, R. Issels, E. Wagner and M. Ogris, Hum. Gene Ther., 2008, 19, 1283–1292 CrossRef CAS PubMed.
- A. Zintchenko, M. Ogris and E. Wagner, Bioconjugate Chem., 2006, 17, 766–772 CrossRef CAS PubMed.
- W. Li, J. Li, J. Gao, B. Li, Y. Xia, Y. Meng, Y. Yu, H. Chen, J. Dai, H. Wang and Y. Guo, Biomaterials, 2011, 32, 3832–3844 CrossRef CAS PubMed.
- M. Nakayama and T. Okano, Macromolecules, 2008, 41, 504–507 CrossRef CAS.
- W. Li, H. Zhao, W. Qian, H. Li, L. Zhang, Z. Ye, G. Zhang, M. Xia, J. Li, J. Gao, B. Li, G. Kou, J. Dai, H. Wang and Y. Guo, Biomaterials, 2012, 33, 5349–5362 CrossRef CAS PubMed.
- W. Li, S. Feng and Y. Guo, Nanomedicine, 2012, 7, 1235–1252 CrossRef CAS PubMed.
- J. M. Lord, L. M. Roberts and J. D. Robertus, FASEB J., 1994, 8, 201–208 CAS.
- P. Li, J. M. Zhu, P. Sunintaboon and F. W. Harris, Langmuir, 2002, 18, 8641–8646 CrossRef.
- J. Zhu, A. Tang, L. P. Law, M. Feng, K. M. Ho, D. K. Lee, F. W. Harris and P. Li, Bioconjugate Chem., 2005, 16, 139–146 CrossRef CAS PubMed.
- W. Li, M. Nakayama, A. Jun and T. Okano, Polymer, 2011, 52, 3783–3790 CrossRef CAS.
- M. Nakayama, T. Okano, T. Miyazaki, F. Kohori, K. Sakai and M. Yokoyama, J. Controlled Release, 2006, 115, 46–56 CrossRef CAS PubMed.
- N. Nishiyama, Y. Bae, K. Miyata, S. Fukushima and K. Kataoka, Drug Discovery Today: Technol., 2005, 2, 21–26 CrossRef CAS PubMed.
- G. Odian, John wiley& sons, Inc., 2004, ch. 4.
- R. G. Gilbert, A Mechanistic Approach, Academic Press, 1995, ch. 2–5 Search PubMed.
- M. Nakayama, Y. Kawahara, J. Akimoto, H. Kanazawa and T. Okano, Colloids Surf., B, 2012, 99, 12–19 CrossRef CAS PubMed.
- E. Ayano, M. Karaki, T. Ishihara, H. Kanazawa and T. Okano, Colloids Surf., B, 2012, 99, 67–73 CrossRef CAS PubMed.
- M. Noga, D. Edinger, R. Kläger, S. V. Wegner, J. P. Spatz, E. Wagner, G. Winter and A. Besheer, Biomaterials, 2013, 34, 2530–2538 CrossRef CAS PubMed.
- M. D. Lavigne, S. S. Pennadam, J. Ellis, L. L. Yates, C. Alexander and D. C. Górecki, J. Gene Med., 2007, 9, 44–54 CrossRef CAS PubMed.
- B. R. Twaites, C. de las Heras Alarcón, D. Cunliffe, M. Lavigne, S. Pennadam, J. R. Smith, D. C. Górecki and C. Alexander, J. Controlled Release, 2004, 97, 551–566 CrossRef CAS PubMed.
- J. S. deVendômois, F. Roullier, D. Cellier and G. E. Séralini, Int. J. Biol. Sci., 2009, 5, 706–726 CrossRef.
- H. Maeda, T. Sawa and T. Konno, J. Controlled Release, 2001, 74, 47–61 CrossRef CAS PubMed.
- D. Mishra, H. C. Kang and Y. H. Bae, Biomaterials, 2011, 32, 3845–3854 CrossRef CAS PubMed.
- W. Li, S. Feng and Y. Guo, Nanomedicine, 2012, 7, 169–172 CrossRef CAS PubMed.
- I. Teraoka, John Wiley Sons, Inc., New York, 2002, ch. 3 Search PubMed.
- A. Tamura, M. Oishi and Y. Nagasaki, J. Controlled Release, 2010, 146, 378–387 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra13707e |
| This journal is © The Royal Society of Chemistry 2016 |