DOI:
10.1039/C6RA13650H
(Paper)
RSC Adv., 2016,
6, 88300-88305
Lipids influence the proton pump activity of photosynthetic protein embedded in nanodiscs†
Received
26th May 2016
, Accepted 4th September 2016
First published on 5th September 2016
Abstract
We report the lipid-composition dependent photocycle kinetics and proton pump activity of bacteriorhodopsin (bR) embedded in nanodiscs composed of different lipids. Using time-resolved spectroscopy and electrochemical methods, we were able to comprehensively understand the kinetics of the photocycle and the corresponding proton pumping activity as the composition of the charged lipids were systematically adjusted. We found that positively-charged lipids assist in repulsing protons from bR, thus increasing the concentration of the non-bounded protons in the bulk. In contrast, the negatively-charged lipids assist in entrapping the protons in the proximity of bR during the photocycle, preserving the electromotive force across the lipid bilayers which is essential for the vitality of the lateral proton transport and the bioenergetics.
1 Introduction
Proton movement is thought to be essential for miscellaneous biological processes, e.g. ATP synthesis.1 Many efforts have been made to show the involvement of lipids in proton movement along the membrane surface in terms of the autoionization of water,2 hydration,3 signs of the surface charges of lipids,4 and anomalous surface diffusion.5 However, little experimental data is available to directly illustrate the dynamical proton transportation process.6,7 In the present work, we applied time-resolved spectroscopy and electrochemical measurements to show the specific roles of lipids in the proton transportation of the photosynthetic protein during the light-induced proton pumping process. Our study will benefit future works utilizing lipids in controlling electromotive force at the lipid bilayers or artificial membrane.
Lateral ion transport in lipid bilayers is essential for maintaining the electromotive force between the cytoplasmic and extracellular sides of bacteriorhodopsin (bR),8–10 a well-known light-driven proton-pump transmembrane protein.11 Upon excitation of bR with a green-yellow photon, photocycle is initiated and the corresponding intermediates are spectrally distinguishable in the visible region, in terms of the status of the retinal Schiff base,12
The completion of the photocycle pumps a proton from the cytoplasmic side to the extracellular side.13 The kinetics of the intermediate M directly correlates to the deprotonation and protonation of the Schiff base, coupled with the proton release to the bulk through a hydrogen bond network at the extracellular proximity, called proton release group.14–17 It is believed that the lipids played critical roles in maintaining the photocycle activity.18,19
In this work, bR served as the light-initiated proton generator to investigate the proton exchange between lipid and bulk water. Bacteriorhodopsins were reconstituted into lipid nanodiscs to mimic the native lipid-bilayer environment, since nanodiscs were ideal due to their great stability, size-tunability, and mono-dispersivity.20–22 The detailed preparation of bR in the lipid nanodiscs followed a recent work by Lee et al.23 The chemical structures of the five chosen lipids, 1,2-dioleoyl-3-trimethyl-ammonium-propane chloride salt (DOTAP), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] sodium salt (DOPG), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) sodium salt (POPG), are shown in Fig. 1. These lipids were either with dioleoyl group or 1-palmitoyl-2-oleoyl group as the hydrophobic tail. Their phase transition temperatures were all below room temperature, allowing us to perform experiments at lipid liquid phase.24–28 One set of bRs was incorporated into nanodiscs assembled with dioleoyl-tail lipids, where different ratios of head groups were used to tune the lipid-bilayer surface charge status. This allowed the comparison of the photocurrent kinetics of bR surrounded by different ratios of charged lipids. We also prepared another set of bRs embedded in nanodiscs made with 1-palmitoyl-2-oleoyl tail lipids to ensure the observed lipid charge dependent photocurrent magnitude is a general trend.
 |
| | Fig. 1 The molecular structures of the lipids: 1,2-dioleoyl-3-trimethyl-ammonium-propane chloride salt (DOTAP, +), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC, +/−), 1,2-dioleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] sodium salt (DOPG, −), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC, +/−) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) sodium salt (POPG, −). | |
2 Materials and methods
2.1 Preparation of bR in lipid nanodiscs
Monomeric bR in Triton X-100 micelle. The detergent solubilized monomeric bR (mbR) was prepared by mixing purified purple membrane (PM) from H. salinarum S9 strain with detergent Triton X-100 at a ratio of 1 to 7 (w/w) (ref. 29) in dark environment for 72 hours at 25 °C. The monomerized bR was collected from the supernatants after three times of centrifugation at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 × g for 30 minutes for further experiments. The absorption maximum of the retinal of mbR in micelle after light adaptation for 30 minutes shifted from 568 to 553 nm.29
400 × g for 30 minutes for further experiments. The absorption maximum of the retinal of mbR in micelle after light adaptation for 30 minutes shifted from 568 to 553 nm.29
Expression of membrane scaffold protein. The plasmid used to express the membrane scaffold protein MSP1E3D1 was obtained from Professor Wagner of Harvard Medical School. MSP1E3D1 was expressed using Escherichia coli Rosetta 2 (DE3) and grown at 37 °C in 1 L of TB medium using a 2 L Fernbach flask. The protein was induced with 1 mM IPTG when OD600 reached above 0.8 for 5 h before harvest, and was then purified using procedures outlined in literature.22,30
Monomeric bR in lipid nanodisc. 1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] sodium salt (DOPG), 1,2-dioleoyl-3-trimethyl-ammonium-propane chloride salt (DOTAP), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) sodium salt (POPG) were purchased from Avanti Polar Lipids and were solubilized using Tris–HCl buffer at pH 7.5 containing 200 mM Triton TX-100 into stock solutions of 50 mM lipid concentration. Different molar ratios of DOPG/DOPC or DOTAP/DOPC mixtures, as well as POPG/POPC mixtures, were made using the stock solutions. The suitable lipid/triton mixture, solubilized monomeric bR and MSP1E3D1 solution were combined with the final molar ratio shown in Table 1. The samples were incubated for 2 hours at 4 °C before detergent was removed by overnight treatment at 4 °C with 800 mg of BioBeads SM-2 (Bio-Rad Laboratories) per 1 ml assembly. The BioBeads were then separated from the assembled nanodisc solution using centrifugation. Fast centrifugation (no. 5424R, Eppendorf) at a top speed of 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm (21
000 rpm (21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 130 × g) was employed to remove additional precipitation before purification using anionic exchange chromatography and size-exclusion chromatography.
130 × g) was employed to remove additional precipitation before purification using anionic exchange chromatography and size-exclusion chromatography.
Table 1 The molar ratios of lipid and bR in different bR embedded lipid nanodiscs
| |
DOTAP/DOPC 10/90 |
DOPG/DOPC 10/90 |
DOPG/DOPC 50/50 |
DOPG/DOPC 100/0 |
| Lipid/MSP |
65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
| MSP/mbR |
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
| |
|
POPG/POPC 10/90 |
POPG/POPC 50/50 |
POPG/POPC 100/0 |
| Lipid/MSP |
|
85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
| MSP/mbR |
|
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
2.2 Characterization of bR in lipid nanodiscs
Anionic exchange and size-exclusion chromatography. bR embedded lipid nanodisc was applied onto an anionic exchange column (Resource Q, GE Healthcare) and eluted with buffer containing 25 mM Tris–HCl, 0.5 mM EDTA at pH 8 and a gradient of linearly increasing salt concentration from 0 to 1 M NaCl. The gel filtration profiles, monitoring the absorption at 280 nm, of nanodiscs samples prepared using different ratios of DOPC, DOPG, and DOTAP are shown in Fig. 2a and the results for the samples prepared with different ratios of POPC and POPG are shown in Fig. S1.† The samples were then collected and condensed for further purification by size exclusion chromatography to part the nanodisc from unwanted aggregation. bR embedded lipid nanodisc solution was applied onto a size exclusion column (Superdex 200 prep grade gel, GE Healthcare) at 4 °C with elution buffer consisting of 25 mM Tris–HCl, 100 mM NaCl and 0.5 mM EDTA at pH 7.5. Fractions expected to contain assembled bR embedded nanodisc were checked with SDS PAGE using 5–13% Tris–glycine gel to check for the presence of bR and MSP1E3D1, in order to verify the incorporation of bR into lipid nanodisc (Fig. 3); (*) was confirmed to be MSP1E3D1(−), MSP1E3D1 with N terminal his tagged removed, cleaved by protease during nanodisc assembly.
 |
| | Fig. 2 (a) Anion-exchange chromatography profiles of bR embedded in nanodisc composed of DOPG/DOPC = 100/0 (blue), DOPG/DOPC = 50/50 (green), DOPG/DOPC = 10/90 (orange) and DOTAP/DOPC = 10/90 (red), using a gradient of linearly increasing NaCl concentration in elution buffer with 25 mM Tris, 0.5 mM EDTA at pH 8. (b) Size exclusion chromatography profile from Superdex 200 10/300 GL of bR embedded in DOPG/DOPC = 10/90 nanodisc. Shown in inset is the MALDI-TOF mass spectrum of combined fraction of 1 to 5. | |
 |
| | Fig. 3 SDS-PAGE of fractions collected during size exclusion chromatography, with bR, MSP1E3D1 and MSP1E3D1(−) standard samples and lanes corresponding to the fraction taken at specific elution volume shown in Fig. 2(b). (*) was confirmed to be MSP1E3D1(−), MSP1E3D1 with N terminal his tagged removed, cleaved by protease during nanodisc assembly. | |
Matrix assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrum. Sample fractions collected from size exclusion were combined and exchanged into distilled water using Vivaspin Concentrator (Sartorius Stedim Biotech), before adding organic solvent mixture of acetone and methanol at a ratio of 7 to 1, respectively. The sample mixture was then rested on ice for 1.5 h before obtaining protein precipitate by fast centrifugation. The protein precipitate was dissolved using trifluoroacetic acid (TFA) and was mixed with DHB matrix containing 50 mg ml−1 2,5-dihydroxybenzoic acid (DHB), 70% acetonitrile (ACN) and 0.1% TFA. The mass spectrum was recorded using Bruker Microflex LRF MALDI mass spectrometer (Bruker Daltonics).
Steady-state spectra. The steady-state ultraviolet-visible (UV-Vis) absorption spectra were monitored with a spectrometer (USB4000-UV-VIS, Ocean Optics) and a cuvette of optical path length of 1 cm was employed in these measurements. The un-normalized spectra are shown in Fig. S2a.†
2.3 Time-resolved absorption of photocycle intermediates
An optical parametric oscillator (basiScan M/120/HE, Spectra-Physics) pumped by a frequency-tripled Nd:YAG laser (INDI-40-10, Spectra-Physics) provided the 6 ns pulses at 532 nm. A mechanical shutter (VS25S2T1, Uniblitz) was utilized to decouple the 10 Hz laser irradiation to 2 Hz for bR embedded DOTAP nanodiscs and 5 Hz for others to avoid over-shooting. The laser flux was controlled at 0.5 mJ cm−2. A deuterium/tungsten halogen lamp (DH-2000, Ocean Optics), attenuated by neutral density filter (Newport) and combined with color filters, served as the stationary probing light. After passing through the cell compartment, the probe beam was further dispersed by a monochromator (Model 218, McPherson) and narrowed by the bandpass filters for the detection of the recovery of parent depletion (560 nm), intermediates M (410 nm) and O (650 nm), respectively. The excitation laser beam was aligned perpendicularly to the probe beam and overlapped in the center of the sample cuvette. The optical modulation was monitored by a photomultiplier (R928, Hamamatsu) and further recorded by a 200 MHz digital oscilloscope (24MXs-B, LeCroy). Temporal profiles upon 1200 laser excitations were averaged for better signal-to-noise ratio. The evolution of the absorbance difference, ΔAt, was derived using the following equation,where St and S0 represent the dc-coupled voltages in the presence and absence of the excitation laser, respectively. The un-weighted temporal profiles are shown in Fig. S2b–d.†
2.4 Time-resolved photoelectric measurements
The construction of the electrochemical cell followed a previous report by Kuo and Chu (ref. 31). The concentration of the bR was 5 μM in the presence of 50 mM NaCl in the working half-cell. The same concentration of NaCl was used in the reference half-cell. 240 μl of the solutions were injected in both half cells. The same pulsed excitation laser in transient absorption measurements was employed to irradiate the electrochemical cell. The laser flux was controlled at 0.5 mJ cm−2 at a repetition rate of 2 Hz to avoid overshooting. The laser beam was introduced via wave-guider (Model 77629, Oriel Instruments) to the front of the electrochemical cell. The photocurrent was amplified by a current amplifier (SR570, Stanford Research System), averaged over 200 laser shots, and recorded by a 400 MHz transient digitizer (44MXs-B, LeCroy).
3 Results and discussions
3.1 Characterization of bR in nanodiscs
The bR-embedded nanodiscs were characterized using anion exchange chromatography and MALDI-TOF mass spectroscopy (Fig. 2). Anion exchange chromatography with a linearly increasing salt gradient (Fig. 2a) showed an increase in negative surface charge of bR embedded nanodisc as the content of the positively-charged lipid DOTAP was decreased and the negatively-charged DOPG was increased. Similar phenomena were observed for nanodisc samples assembled with POPC and POPG, as shown in Fig. S1.† Size exclusion chromatography (Fig. 2b) was employed to purify the sample based on size. The symmetrical peak was verified to contain bR embedded nanodisc, as both bR and membrane scaffold protein (MSP) were shown on SDS-PAGE (Fig. 3) and mass spectrum (inset of Fig. 2b). The kinetics of bR photocycle and the proton relay process were characterized using transient absorption and electrochemical method,31–33 respectively. Beer's law states that the absorbance is proportional to the concentration, thus the transient absorption technique was employed to illustrate the concentration evolution of photocycle intermediates, characterized at the given wavelengths. Moreover, the electrochemical approach was employed to illustrate the proton concentration alteration in the aqueous solution during the photocycle.31–33
When the bRs embedded in the nanodiscs in the presence of different lipids were light adapted, the absorption contours attributed to the protonated retinal Schiff base were approximated at 560 nm (Fig. 4a and 5a). It should be noted that no additional band at 390 nm formed, indicating that the hydrolysis of retinal34 did not take place in different lipid environments. Moreover, reducing the content of negatively-charged lipids led to the blue-shifted absorption band of the protonated retinal Schiff base. It could be attributed to the alteration of the population of the all-trans, 15-anti and the 13-cis, 15-syn retinal in bR interior after the light adaptation in different lipid compositions. Scherrer et al. reported a 13 nm blue-shift from the all-trans, 15-anti retinal to the 13-cis, 15-syn.35 Thus, the observed blue-shift in the absorption contour suggested that a decrease in the negatively-charged lipid content resulted in an increase in population of the 13-cis retinal.
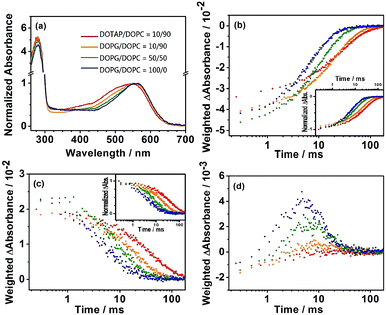 |
| | Fig. 4 (a) Normalized steady-state absorption spectra of bR embedded in nanodiscs composed of DOPG/DOPC = 100/0 (blue), DOPG/DOPC = 50/50 (green), DOPG/DOPC = 10/90 (orange) and DOTAP/DOPC = 10/90 (red) in the presence of 50 mM NaCl. The pH was controlled at 7.03 ± 0.01 by adding NaOH or HCl without using buffer solution. The temporal profiles of (b) the recovery of the parent state at 560 nm, (c) the intermediate M at 410 nm, and (d) the intermediate O at 650 nm upon 532 nm excitation (0.5 mJ cm−2 per pulse) of these samples. The temporal profiles were weighted by their corresponding steady-state optical density at 532 nm. The un-normalized absorption spectra are supplemented in ESI.† The normalized traces of frames (b) and (c) were shown in the insets for comparison. | |
 |
| | Fig. 5 (a) Normalized steady-state absorption spectra of bR embedded in nanodiscs composed of POPG/POPC = 100/0 (blue), POPG/POPC = 50/50 (green) and POPG/POPC = 10/90 (orange) in the presence of 50 mM NaCl. The pH was controlled at 7.03 ± 0.01 by adding NaOH or HCl without using buffer solution. The temporal profiles of (b) the recovery of the parent state at 560 nm, (c) the intermediate M at 410 nm, and (d) the intermediate O at 650 nm upon 532 nm excitation (0.5 mJ cm−2 per pulse) of these samples. The temporal profiles were weighted by their corresponding steady-state optical density at 532 nm. The un-normalized absorption spectra are supplemented in ESI.† The normalized traces of frames (b) and (c) were shown in the insets for comparison. | |
3.2 Kinetics of photocycle intermediates
The evolutions of the recovery of parent depletion (Fig. 4b and 5b), the intermediate M at 410 nm (Fig. 4c and 5c), and the intermediate O at 650 nm (Fig. 4d and 5d), altered significantly as the content of differently charged lipids changed and was observed for both types of hydrophobic tails. The weighted absorbance difference denoted the observed temporal profiles divided by the optical density of the steady-state absorption spectra at 532 nm of the samples to eliminate the slight deviation of the sample concentration. Previous studies have shown that the retinal in 13-cis configuration is not capable of undergoing the conventional proton pump photocycle upon photoexcitation,36,37 therefore the instantaneous depletion of the parent state upon photoexcitation changes in different lipid compositions is due to the difference of all-trans retinal population. Because the excitation power of the 532 nm laser was kept the same, the instantaneous depletion of bR parent state explicitly indicated the percentile of the photo-excitable bR. The sample of bR embedded in DOTAP/DOPC = 10/90 nanodisc exhibited the weakest depletion (Fig. 4b) in the DO series. This was consistent with the steady-state absorption results that the effective concentration of bR with photoactive all-trans retinal was reduced as the content of positively-charged lipid was increased. The transient population of the intermediate M (Fig. 4c) at the early period also exhibited similar trends. Moreover, the normalized profiles in Fig. 4b and c indicated that as the content of the negatively-charged lipids decreased, the kinetics of parent recovery and intermediate M were decelerated. Concomitantly, the transient population of intermediate O (Fig. 4d) also diminished as the content of the positively-charged lipids increased. For the nanodisc samples made with the mixtures of POPG and POPC, a trend similar to mixtures containing the DOPG and DOPC was observed, as shown in Fig. 5b–d. The present works agreed with a recent study.23
3.3 Photocurrent of bR in nanodiscs
The temporal profiles of the photocurrents of DO and PO series upon pulsed excitation are shown in Fig. 6a and b, respectively. The waveforms were similar but the magnitudes were reduced as the content of PG moiety increased. Comparing with the depletion of the parent state at the early state of photoexcitation in DO series, the photocycle of bR in DOTAP/DOPC = 10/90 was expected to proceed in the weakest fashion. However, the photocurrent magnitude of bR in DOTAP/DOPC = 10/90 nanodisc was shown to be greater than bR in DOPG containing nanodiscs. This could be due to the intrinsic properties of the hydrophilic heads of the lipids. When the photocycle is initiated, the proton relay starts from the protonated Schiff base to the proton release group at extracellular proximity. If there are more negatively-charged functional groups, such as phosphatidylglycerol in DOPG and POPG, in the vicinity of the proton release group, the proton could be trapped at the surface of these lipids (Fig. 7a). However, as the hydrophilic heads of the lipids become positively-charged or less negatively-charged, the attraction to protons is weakened and the positive charge assists in repulsing the proton from bR, thus increasing the concentration of the non-bounded protons in the bulk solution and the photocurrent amplitude (Fig. 7b). Comparing with the transient absorption (Fig. 4b–d and 5b–d), we demonstrated that the protons retained in the lipid bilayers by the negatively-charged lipids assist in completing the photocycle and shortening the lifetime of the photocycle, implying that the duty cycles can be increased in a given photo-exposure. This conclusion is supported by the fact that the natural lipid composition of a primitive unit of purple membrane, containing trimeric bR and surrounding lipids,18,19,38 is mainly negatively charged, such as glycolipid sulfate, phosphatidylglycerophosphate methyl ester, or phosphatidylglycerol.39 In addition, the change in the hydrophobic tails did not alter the trend that the increase in the content of the negatively-charged lipids reduced the magnitude of the photocurrent. This suggests that the phenomenon was dominated by the charge of the hydrophilic head. While we showed that the sign of the charged hydrophilic heads is essential for the alteration in the bR photocycle and proton pump kinetics, the slightly different photocurrents in lipids with dioleoyl and 1-palmitoyl-2-oleoyl tails, respectively, could be observed. This difference could potentially result from other minor factors such as different lipid packing orders and different phase transition temperatures.
 |
| | Fig. 6 The temporal profiles of the photocurrent upon 532 nm pulsed excitation (0.5 mJ cm−2 per pulse) of the samples in Fig. 4 and Fig. 5. The traces were weighted by their corresponding steady-state optical density at 532 nm. | |
 |
| | Fig. 7 The schematics of the proton relay for bR embedded in (a) negatively-charged and (b) positively-charged lipids. | |
4 Conclusions
The purpose of this work was to unravel the roles of lipids in influencing the electromotive force generated by the light-driven proton pump of bacteriorhodopsin in lipid nanodiscs and to provide an experimental strategy to controllably utilize this chemical potential. Using the time-resolved spectroscopic and electrochemical approaches, we were able to comprehensively understand the kinetics of the photocycle and the corresponding proton pumping activity as the composition of the charged lipids were systematically adjusted. We found that positively-charged lipids assist in repulsing protons from bR, thus increasing the concentration of the non-bounded protons in the bulk. In contrast, the negatively-charged lipids assist in entrapping the protons in the proximity of bR during the photocycle, preserving the electromotive force across the lipid bilayers which is essential for bioenergetics.
Acknowledgements
This work is supported by the Ministry of Science and Technology of Taiwan (MOST 103-2113-M-007-010-MY2, MOST 103-2113-M-001-001-MY2). Y. Hsin was supported by the scholarship of College Student Participation in Research Projects from the Ministry of Science and Technology of Taiwan (104-2815-C-007-083-M). In addition, T. Y. Yu and V. Yeh would like to acknowledge the support from Academia Sinica Nano Program.
References
- P. Mitchell, Nature, 1961, 191, 144 CrossRef CAS PubMed
 .
. - A. Mashaghi, P. Partovi-Azar, T. Jadidi, M. Anvari, S. P. Jand, N. Nafari, M. R. R. Tabar, P. Maass, H. J. Bakker and M. Bonn, J. Phys. Chem. C, 2013, 117, 510 CAS
 .
. - A. Mashaghi, P. Partovi-Azar, T. Jadidi, N. Nafari, P. Maass, M. R. R. Tabar, M. Bonn and H. J. Bakker, J. Chem. Phys., 2012, 136, 114709 CrossRef PubMed
 .
. - T. Yamashita and G. A. Voth, J. Phys. Chem. B, 2010, 114, 592 CrossRef CAS PubMed
 .
. - M. G. Wolf, H. Grubmüller and G. Groenhof, Biophys. J., 2014, 107, 76 CrossRef CAS PubMed
 .
. - T. Sandén, L. Salomonsson, P. Brzezinski and J. Widengren, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 4129 CrossRef PubMed
 .
. - M. Brändén, T. Sandén, P. Brzezinski and J. Widengren, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 19766 CrossRef PubMed
 .
. - J. Heberle, J. Riesle, G. Thiedemann, D. Oesterhelt and N. A. Dencher, Nature, 1994, 370, 379 CrossRef CAS PubMed
 .
. - P. Scherrer, U. Alexiev, T. Marti, H. G. Khorana and M. P. Heyn, Biochemistry, 1994, 33, 13684 CrossRef CAS PubMed
 .
. - U. Alexiev, R. Mollaaghababa, P. Scherrer, H. G. Khorana and M. P. Heyn, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 372 CrossRef CAS
 .
. - D. Oesterhelt and W. Stoeckenius, Nat. New Biol., 1971, 233, 149 CrossRef CAS PubMed
 .
. - J. K. Lanyi, J. Phys. Chem. B, 2000, 104, 11441 CrossRef CAS
 .
. - R. A. Mathies, S. W. Lin, J. B. Ames and W. T. Pollard, Annu. Rev. Biophys. Biophys. Chem., 1991, 20, 491 CrossRef CAS PubMed
 .
. - K. J. Rothschild, J. Bioenerg. Biomembr., 1992, 24, 147 CrossRef CAS PubMed
 .
. - G. Váró and J. K. Lanyi, Biochemistry, 1991, 30, 5008 CrossRef
 .
. - P. Phatak, N. Ghosh, H. Yu, Q. Cui and M. Elstner, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 19672 CrossRef CAS PubMed
 .
. - L. Zimányi, G. Váró, M. Chang, B. Ni, R. Needleman and J. K. Lanyi, Biochemistry, 1992, 31, 8535 CrossRef
 .
. - S. Dracheva, S. Bose and R. W. Hendler, FEBS Lett., 1996, 382, 209 CrossRef CAS PubMed
 .
. - M. K. Joshi, S. Dracheva, A. K. Mukhopadhyay, S. Bose and R. W. Hendler, Biochemistry, 1998, 37, 14463 CrossRef CAS PubMed
 .
. - F. Hagn, M. Etzkorn, T. Raschle and G. Wagner, J. Am. Chem. Soc., 2013, 135, 1919 CrossRef CAS PubMed
 .
. - T. H. Bayburt and S. G. Sligar, FEBS Lett., 2010, 584, 1721 CrossRef CAS PubMed
 .
. - I. G. Denisov, Y. V. Grinkova, A. A. Lazarides and S. G. Sligar, J. Am. Chem. Soc., 2004, 126, 3477 CrossRef CAS PubMed
 .
. - T.-Y. Lee, V. Yeh, J. Chuang, J. C. C. Chan, L.-K. Chu and T.-Y. Yu, Biophys. J., 2015, 109, 1899 CrossRef CAS PubMed
 .
. - A. E. Regelin, S. Fankhaenel, L. Gürtesch, C. Prinz, G. von Kiedrowski and U. Massing, Biochim. Biophys. Acta, 2000, 1464, 151 CrossRef CAS
 .
. - E. J. Findlay and P. G. Barton, Biochemistry, 1978, 17, 2400 CrossRef CAS PubMed
 .
. - D. Casey, K. Charalambous, A. Gee, R. V. Law and O. Ces, J. R. Soc., Interface, 2014, 11, 20131062 CrossRef PubMed
 .
. - T. Wiedmann, A. Salmon and V. Wong, Biochim. Biophys. Acta, 1993, 1167, 114 CrossRef CAS
 .
. - N. Shimokawa, M. Nagata and M. Takagi, Phys. Chem. Chem. Phys., 2015, 17, 20882 RSC
 .
. - J. Wang, S. Link, C. D. Heyes and M. A. El-Sayed, Biophys. J., 2002, 83, 1557 CrossRef CAS PubMed
 .
. - T. H. Bayburt, Y. V. Grinkova and S. G. Sligar, Nano Lett., 2002, 2, 853 CrossRef CAS
 .
. - C.-L. Kuo and L.-K. Chu, Bioelectrochemistry, 2014, 99, 1 CrossRef CAS PubMed
 .
. - K. Koyama, N. Yamaguchi and T. Miyasaka, Science, 1994, 265, 762 CAS
 .
. - L.-K. Chu, C.-W. Yen and M. A. El-Sayed, Biosens. Bioelectron., 2010, 26, 620 CrossRef CAS PubMed
 .
. - J. P. Schlebach, Z. Cao, J. U. Bowie and C. Park, Protein Sci., 2012, 21, 97 CrossRef CAS PubMed
 .
. - P. Scherrer, M. K. Mathew, W. Sperling and W. Stoeckenius, Biochemistry, 1989, 28, 829 CrossRef CAS PubMed
 .
. - I. Logunov, W. Humphrey, K. Schulten and M. Sheves, Biophys. J., 1995, 68, 1270 CrossRef CAS PubMed
 .
. - N. Mizuide, M. Shibata, N. Friedman, M. Sheves, M. Belenky, J. Herzfeld and H. Kandori, Biochemistry, 2006, 45, 10674 CrossRef CAS PubMed
 .
. - J. K. Lanyi, Mol. Membr. Biol., 2004, 21, 143 CrossRef CAS PubMed
 .
. - A. Corcelli, V. M. T. Lattanzio, G. Mascolo, P. Papadia and F. Fanizzi, J. Lipid Res., 2002, 43, 132 CAS
 .
.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details and additional data. See DOI: 10.1039/c6ra13650h |
| ‡ These authors equally contributed to this work. |
|
| This journal is © The Royal Society of Chemistry 2016 |
Click here to see how this site uses Cookies. View our privacy policy here. ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 × g for 30 minutes for further experiments. The absorption maximum of the retinal of mbR in micelle after light adaptation for 30 minutes shifted from 568 to 553 nm.29
400 × g for 30 minutes for further experiments. The absorption maximum of the retinal of mbR in micelle after light adaptation for 30 minutes shifted from 568 to 553 nm.29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm (21
000 rpm (21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 130 × g) was employed to remove additional precipitation before purification using anionic exchange chromatography and size-exclusion chromatography.
130 × g) was employed to remove additional precipitation before purification using anionic exchange chromatography and size-exclusion chromatography.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1




.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.



