Hollow silica bubble based immobilized trypsin for highly efficient proteome digestion and buoyant separation†
Fenglong Jiaoab,
Rui Zhaib,
Junjie Huangb,
Yukui Zhangc,
Yangjun Zhang*b and
Xiaohong Qian*b
aSchool of Life Science and Technology, Beijing Institute of Technology, Beijing 100081, China
bState Key Laboratory of Proteomics, National Center for Protein Science Beijing, Beijing Institute of Radiation Medicine, Beijing 102206, China. E-mail: 13911734119@163.com; 13683167093@163.com
cNational Chromatographic Research and Analysis Center, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116011, China
First published on 19th August 2016
Abstract
Tryptic digestion before identification and quantification by mass spectrometry is an indispensable process for most proteomics studies. Conventional in-solution digestion process always suffers from incomplete digestion and is time-consuming and expensive. Development of a novel proteolytic digestion method with fast, high efficiency and reusability of the enzyme is still an urgent task. In this study, new hollow silica bubbles-based immobilized trypsin (trypsin@SiB) was developed via brushes structured glycidyl methacrylate (GMA) grafted on the silica bubbles (SiB) by the reversible addition–fragmentation chain transfer polymerization (RAFT) technique followed by trypsin's immobilization on the microsphere. The new immobilized trypsin not only showed high efficiency and stability but also reusability. Due to the low density (0.6 g cm−3) of SiB, it can float over the solution for several minutes so that the immobilized trypsin can be separated and recovered from the system easily. Highly efficient digestion of BSA was achieved with this trypsin@SiB within 1 min and the obtained sequence coverage (92%) was better than that from conventional in-solution digestion for more than 12 h (76%). To further confirm the efficiency of trypsin@SiB for complex proteomic analysis, the protein extracted from human urine was analyzed as a real sample. Within 10 min digestion, 510 protein groups were identified with the LC-MS/MS analysis and database searching, whereas the number of identified proteins after 12 h in-solution digestion was 493 with the same identification conditions. The successful application of trypsin@SiB demonstrated its potential as a high efficient digestion method for future proteomics analysis.
1. Introduction
Proteomics has attracted considerable attention over last couple of decades because of the urgent demand for identification of proteins that play very important roles in all biological processes1–3 on a large scale. The main research strategies of proteomics are “top-down”, “middle-down” and “bottom-up”. In the strategy of “middle-down” and “bottom-up”, proteins are always digested to peptides, followed by liquid chromatography-mass spectrometry (LC-MS) analysis for protein identification and quantification.4–7 Peptides, due to their lower molecular weights, may improve the sensitivity of identification by mass spectrometry and meet the mass range of most mass spectrometer with quadrupoles. Therefore, rapid and complete protein digestion is vital to in-depth proteome profiling. However, conventional in-solution digestion suffers from several challenges such as complicated pretreatment steps, time-consuming and autolysis of the enzyme.8 Recently, enzymes immobilized on a solid support has been widely used since high enzyme concentration in limited space can be achieved in this process, leading to short digestion time, low risk for autolysis and reusability of the enzyme.9–12 To date, enzymes have been immobilized on various supports, including graphene-oxide (GO),13,14 hybrid aerogels,15 magnetic nanoparticles,16–18 capillary columns,19,20 nanotubes21,22 and porous reactors.23,24 Although the digestion time has been reduced to several minutes by using these immobilized enzymes with different supports, the separation procedures usually required magnetic force or high-speed centrifugation, which might lead to both time consuming and agglomeration of nano-materials. Therefore, the development of a new enzyme support for easy operation has still attracted great attention in proteomics field.In recent years, hollow silica bubble, as a new class of micro materials, has aroused much interest due to its special physical properties such as low density, refractive index and pore volume.25,26 It has been modified with gold nanoparticles (AuNPs) and served as a convenient platform for the analysis of surface-enhanced Raman scattering (SERS).27,28 Compared with traditional SERS substrate, this Au@SiB is able to concentrate analytes from a solution so that the detection sensitivity is improved. In addition, the new class material has been utilized as functional biomaterials. For example, in glycomics study, it was modified with mercaptophenylboronic (MPB) acid for the enrichment of glycopeptides and separated and recovered for reuse by flotage. The successfully functionalized SiBs displayed an excellent property for the enrichment of glycopeptides due to the highly efficient buoyant separation and specific recognition of MPB to glycopeptides.29
Recently, polymer brushes grafted on the nanoparticles have been widely used in polymerization materials synthesis since it could increase the number of functional groups and greatly improve their interfacial properties. These brushes grafted on nanoparticles were usually used for biological applications such as glycopeptide enrichment, protein-specific Raman imaging and drug delivery.30–33 The main methods for brushes formation are atomic transfer radical polymerization (ATRP) and reversible addition–fragmentation chain transfer polymerization (RAFT).34–36 Since the reaction condition is mild and metal catalysts are not brought into the reaction, RAFT continues to attract researcher's attention.37
In this study, we developed a hollow silica bubbles based immobilized trypsin for high efficient proteome digestion and buoyant separation. Trypsin was immobilized on the brush structure grafted by reversible addition–fragmentation chain transfer polymerization (RAFT). The glycidyl methacrylate (GMA) brushes could not only provide a large capacity for immobilizing enzyme but also affect hydrophobicity of the microsphere and their affinity toward protein substrates.38,39 Because the silica matrix could carry a lot of enzymes and perform an excellent dispersity in both organic and aqueous solutions, the digestion time was dramatically reduced to 1 min. Moreover, the buoyant separation makes the manipulation process convenient. The digestion efficiency was proved to be better than the traditional solution-digestion. Highly efficient digestion of BSA was achieved with this trypsin@SiB within 1 min and the obtained sequence coverage (92%) was better than that from conventional in-solution digestion for more than 12 h (76%). The activity of immobilized enzyme was maintained at its initial level after five repeated experiments due to the excellent dispersity, which might lead to the reduction of interaction of enzymes to a great extent. The performance of protein digestion was further evaluated by the proteins extracted from human urine. The results indicated that the trypsin–GMA@SiB microspheres exhibited better performance compared with conventional in-solution digestion.
2. Materials and methods
2.1 Reagents
Bovine serum album (BSA), glycidyl methacrylate (GMA), trypsin from bovine pancreas, 4-cyano-4-(phenylcarbonothioylthio)pentanoic acid (CPCP), 2,2-azobis(2-methylpropionitrile) (AIBN), sodium periodate (NaIO4), sulfuric acid (H2SO4), DL-dithiothreitol (DTT), iodoacetamide (IAA), 3-aminopropyl-triethoxysilane (APTES), trifluoroacetic acid (TFA), ammonium bicarbonate (NH4HCO3), and α-cyano-4-hydroxycinnamic acid (CHCA) were purchased from Sigma (St. Louis, MO, USA), the reagents 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) were purchased from Alfa Aesar (Ward Hill, MA, USA). S60HS silica bubbles (30 μm, 0.6 g cm−3) were purchased from 3M Company (USA). Ultrapure water was prepared from a Millipore purification system (Billerica, MA, USA).2.2 Preparation of poly-GMA modified SiB and immobilization of trypsin
The amino-modified silica bubbles (NH2@SiB) were synthesized according to the previously reported method.28 Briefly, 200 mg silica bubbles were first activated with 20 mL 5 M H2SO4, and then silanized in an ethanol solution of 10% APTES overnight. The obtained NH2@SiB was washed with ethanol and water three times and dried for further use.Poly-GMA@SiB was synthesized via a reversible addition–fragmentation chain transfer (RAFT) polymerization. 15 mg 4-cyano-4-(phenylcarbonothioylthio)pentanoic acid (CPCP), 11 mg 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC) and 5.8 mg N-hydroxysuccinimide (NHS) were dissolved in 7.5 mL ethanol and the mixture was incubated with shaking for 3 h. Then, NH2@SiB (100 mg) was added into the solution, followed by 18 h reaction at room temperature. After the bubbles rose to the top of the solution, the solution was removed by a syringe. The prepared CPCP-modified SiB was washed with ethanol and water three times and dried at room temperature overnight by vacuum. CPCP@SiB (100 mg), GMA (1 mL), AIBN (5 mg) and dry DMF (10 mL) were added into a Schlenk tube and followed by sonication. The tube was subjected to a liquid nitrogen bath and bubbling nitrogen to remove air. The reaction was performed at 60 °C for 24 h with magnetic stirring. The polymerization was terminated by cooling to room temperature and exposing to air. After the reaction, excess reagents were removed by repeated washing with DMF.
The epoxy groups at the terminal of GMA were converted to aldehyde groups by 0.5 M sulfuric acid at 50 °C for 4 h and 20 mM sodium periodate in the dark at room temperature for 2 h. After washing with pure water for 3 times, the obtained SiBs were dispersed in 10 mL PBS (pH = 7.4) solution with 1 mg mL−1 trypsin. The mixture was kept on a shaker overnight at 4 °C. Finally, trypsin immobilized on SiBs was washed with pure water three times and stored at −20 °C until further use.
2.3 Protein digestion by immobilized trypsin or free trypsin
The size and morphology of the silica bubbles were characterized by S-4800 cold field emission scanning electron microscopy (Hitachi). Fourier transform infrared spectroscopy (FT-IR) characterization was performed on a Fourier spectrophotometer using KBr pellets (Nicolet). Thermogravimetric analysis (TGA) was performed on a Pyris 1 TGA instrument (PerkinElmer). Both SiB and trypsin@SiB were dried at 60 °C prior to each TGA measurement to remove the solution attached to the surface. The samples were heated from 20 °C to 700 °C with a heating rate of 10 °C min−1 under a flow of nitrogen.2.4 Tryptic digests of standard proteins
BSA was used as a standard protein to evaluate the digestion efficiency. For free trypsin digestion, 1 mg BSA was dissolved in 1 mL ammonium bicarbonate solution (50 mM, pH = 8.0) and followed by boiling for 10 min. After that, denatured BSA was reduced with 20 mM DTT at 56 °C for 1 h and alkylated by 7.2 mg IAA at room temperature in the dark for 1 h. The solution was incubated with trypsin at an enzyme–protein ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 (w/w) at 37 °C for 16 h. Free tryptic digests were stored at −20 °C for further use. For immobilized trypsin digestion, the denatured protein solution (1 mg mL−1) was first reduced by DTT and alkylated by IAA. Then, 10 μL slurry of trypsin@SiB was added and vortexed at 37 °C for 1 min. After digestion, the solution was made to stand for several minutes so that trypsin@SiB would float to the surface and can be separated easily. The solution was removed with a syringe for mass spectrometry analysis.
50 (w/w) at 37 °C for 16 h. Free tryptic digests were stored at −20 °C for further use. For immobilized trypsin digestion, the denatured protein solution (1 mg mL−1) was first reduced by DTT and alkylated by IAA. Then, 10 μL slurry of trypsin@SiB was added and vortexed at 37 °C for 1 min. After digestion, the solution was made to stand for several minutes so that trypsin@SiB would float to the surface and can be separated easily. The solution was removed with a syringe for mass spectrometry analysis.
Human urine proteins were precipitated by ice cooled acetone and then dissolved in 100 mM Tris–HCl (pH = 8.5) containing 8 M urea to a concentration of 1 mg mL−1. After reduction by 10 mM DTT at 37 °C for 4 h and alkylated by 20 mM IAA at room temperature in dark for 1 h, the solution was diluted with Tris–HCl to reduce the urea concentration to 1 M. For solution digestion, free trypsin was added into the protein solution at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, and the solution was incubated at 37 °C overnight. For immobilized trypsin digestion, the protein solution was mixed with 5 mg trypsin@SiB and vortexed at 37 °C for 1 min. The digested peptides of human urine proteins were collected by a syringe from the digestion solution after the trypsin@SiB float to the surface of the solution and then desalt with a C18 SPE cartridge. The eluent was collected for mass spectrometry analysis.
50, and the solution was incubated at 37 °C overnight. For immobilized trypsin digestion, the protein solution was mixed with 5 mg trypsin@SiB and vortexed at 37 °C for 1 min. The digested peptides of human urine proteins were collected by a syringe from the digestion solution after the trypsin@SiB float to the surface of the solution and then desalt with a C18 SPE cartridge. The eluent was collected for mass spectrometry analysis.
2.5 MS analysis and database searching
The MALDI-TOF experiments were carried out on an AB Sciex 4800 MALDI-TOF/TOF mass spectrometer (AB Sciex, CA) equipped with a pulsed Nd/YAG laser at 355 nm. 1 μL of a BSA digest and 1 μL of CHCA (7 mg mL−1, 0.1% TFA in 60% CH3CN solution) matrix solution was dropped onto the MALDI plate for MS analysis. Peptide mass fingerprint was searched against a BSA database using MASCOT (version 2.3.01, Matrix Science). Trypsin was chosen as the enzyme and two missed cleavages were allowed. 20 ppm was set as mass tolerance of precursor ion. Carbamidomethyl (Cys) was set as the fixed modification and oxidation (Met) was set as the variable modification.The digested peptide of human urine proteins were resuspended in 10 μL of 0.1% TFA solution and the LC-MS/MS analysis was carried out using a Dionex Ultimate 3000 Nano LC system coupled with a Q-Exactive HF mass spectrometer (Thermo Fisher Scientific, USA) with an ESI nanospray source. 0.1% FA in water and 0.1% FA in ACN were prepared as mobile phase A and B, respectively. The total flow rate was 600 nL min−1 and a 75 min gradient was set as follows: from 6% to 9% buffer B for 8 min, from 9% to 14% buffer B for 16 min, from 14% to 30% buffer B for 36 min, from 30% to 40% buffer B for 15 min and from 40% to 95% buffer B for 3 min. After eluting with 95% buffer for 7 min, the separation system was equilibrated by 6% buffer B for 5 min. The spray voltage was set at 2.0 kV. All MS and MS/MS spectra were acquired in data-dependent acquisition mode and the full mass scan was acquired from m/z 300 to 1400 with a resolution of 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000.
000.
All the LC-MS/MS raw data acquired were submitted to Protein Discoverer software (version 1.4.1.14, Thermo Scientific) for database searching against a human protein database. Trypsin was selected as an enzyme and up to two missed cleavages were allowed. Cysteine residues were set as static modification and oxidation of methionine was set as the variable modification. The mass tolerance of the precursor was 15 ppm and the peptide false discovery rate (FDR) was controlled ≦1%.
2.6 Live subject statement
The authors state that all experiments were performed in compliance with the relevant laws and institutional guidelines. The institutional committee of the National Center for Protein Science Beijing has approved the experiments with live subjects. The authors also state that informed consent was obtained for any experimentation with human subjects and National Center for Protein Science Beijing is committed to the protection and safety of human subjects involved in research.3. Results and discussion
3.1 Preparation and characterization of the trypsin-immobilized SiB
The procedure for the preparation of trypsin-immobilized SiB is shown in Scheme 1. First, SiBs were silanized with 3-aminopropyltriethoxysilane (APTES) to form an amine group packaged formation (SiB–NH2). The chain transfer agent 4-cyano-4-(phenylcarbonothioylthio)pentanoic acid (CPCP) was activated and reacted with NHS, and then attached to the amine groups of SiB–NH2 to form an amide bond. The RAFT technique was utilized for the synthesis of the pGMA SiBs to generate polymer brushes in a controlled condition. Polymerization was carried out with GMA as monomers. The brushes structured GMA on the surfaces of the SiBs provided large numbers of epoxy groups, which could be easily converted to aldehyde groups for enzyme immobilization. Then, trypsin was immobilized on the SiBs by the interaction of amino groups and aldehyde groups, as shown in Scheme 1a. Finally, the prepared trypsin@SiB was utilized for further protein digestion and MS analysis, as shown in Scheme 1b. | ||
| Scheme 1 (a) Schematic of the synthetic process of trypsin@SiB (b) the digestion process of BSA, buoyant separation of trypsin@SiB and MS analysis. | ||
The scanning electron microscopy (SEM) images, as shown in Fig. 1, showed that the SiB nanoparticles had a smooth surface and are spherical in shape with an average diameter of 30 μm (Fig. 1a and c). As shown in Fig. 1b and d, the rough surfaces after reaction indicated that trypsin was immobilized on the microsphere surface successfully.
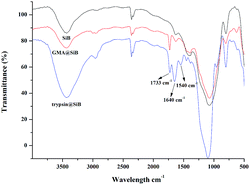
FT-IR was further used to characterize trypsin@SiB. As shown in Fig. 2, the peaks at 1080 cm−1 were assigned to the Si–O–Si and the broad absorption at 3450 cm−1 was attributed to the characteristic absorption of hydroxyl group. The peak at 1720 cm−1 was due to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond, indicating the presence of GMA. After the immobilization of trypsin, two new characteristic peaks at 1640 cm−1 (–CONH amide band I) and 1550 cm−1 (–NH amide band II) appeared,12 indicating the presence of trypsin on the functional surfaces.
O bond, indicating the presence of GMA. After the immobilization of trypsin, two new characteristic peaks at 1640 cm−1 (–CONH amide band I) and 1550 cm−1 (–NH amide band II) appeared,12 indicating the presence of trypsin on the functional surfaces.
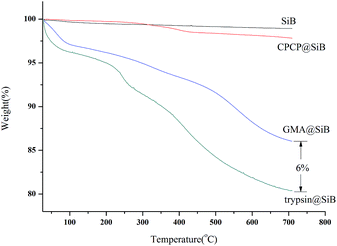
The loading amount of trypsin on SiB was evaluated by measuring the concentration variation of trypsin after reaction, and the result indicated that about 60 μg mg−1 of trypsin was loaded on SiB. Thermogravimetric analysis (TGA) was also performed to confirm the immobilization of trypsin. As shown in Fig. 3, the TGA curves showed that the weight loss of trypsin@SiB was 11%, which indicated the presence of a high amount of trypsin immobilized on the microsphere surface.
3.2 Digestion of standard BSA by trypsin@SiB
BSA was used as a standard protein to evaluate the digestion performance of trypsin immobilized on the silica bubbles. The digestion time was first tested using BSA solution with a concentration of 400 μg mL−1 (1 mL ammonium bicarbonate solution containing 400 μg BSA, pH = 8.3). 5 mg SiB and trypsin@SiB were incubated with 100 μL BSA solution (0.1 μg μL−1) for 1 min. Then, 10 μL of BSA solution, BSA solution digested by SiB and trypsin@SiB were collected for SDS-PAGE analysis. As shown in Fig. 4, in lane 2, the BSA concentration treated with SiB did not change compared with standard BSA, whereas the BSA could not be detected by SDS-PAGE analysis after 1 minute digestion with trypsin@SiB was performed, as shown in lane 3. The result demonstrated that trypsin has been immobilized on the SiB and high efficient digestion was obtained within 1 min, which might be contributed to the increased enzyme content. | ||
| Fig. 4 SDS-PAGE analysis of digested BSA (lane 1: BSA; lane 2: BSA digested by SiB; lane 3: BSA digested by trypsin@SiB). | ||
MALDI-TOF-MS experiments were carried out to evaluate the digestion efficiency. As shown in Fig. 5, the MALDI-TOF-MS spectra of BSA digested by trypsin@SiB for 1 min exhibited more peaks and high sequence coverage (92%) than that of free trypsin digestion for 12 h (76%) in a single experiment. The specific results are shown in Table S1 and S2.† Moreover, no obvious peaks could be detected exceeding 3000 m/z in both the spectra, which indicated that the digestion by trypsin@SiB was completed by free trypsin in a much shorter time, and the average sequence coverage of the identified peptides by either trypsin@SiB digestion or free trypsin digestion was evaluated and the results showed that an average sequence coverage 85% was obtained by trypsin@SiB digestion for BSA in 1 minute incubation, which was significantly higher than that of conventional in-solution digestion (73%) overnight, as shown in Fig. 6.
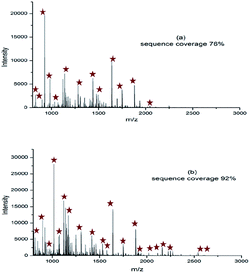 | ||
| Fig. 5 MALDI-TOF-MS spectra of BSA digested by (a) free trypsin for 12 h, (b) trypsin@SiB for 1 min. | ||
To evaluate the reusability, trypsin@SiB was used for digestions of BSA for 7 times. After each digestion, the solution was removed by a syringe and trypsin@SiB was recovered and washed by water for 3 times before next use. The average sequence coverage was 79%. The sequence coverage in 7 runs (81%, 83%, 78%, 77%, 79%, 77%, and 80%) indicated that trypsin@SiB had a good reusability and high digestion efficiency. The stability of trypsin@SiB was tested after storing at −20 °C for 1 month and the result was shown in Table 1; a high enzymatic activity was still maintained with a sequence coverage of BSA 84%, which was higher than that obtained by in-solution digestion.
| Enzymatic method | Trypsin@SiB after storing at −20 °C for 1 month | In-solution digestion |
|---|---|---|
| Digestion time | 1 min | 12 h |
| Sequence coverage (peptides matched) | 84% (59), 81% (55), 87% (64) | 77% (51), 76% (54), 76% (51) |
| Average sequence coverage | 84% | 76% |
4. Conclusions
In this study, a novel silica bubbles-based immobilized trypsin was developed to facilitate the enzymatic digestion of proteins. High trypsin digestion efficiency was achieved due to the large amount of trypsin immobilized on SiB. The sequence coverage obtained with trypsin@SiB in 1 min was higher than that of in-solution digestion overnight. The urine proteome study by LC-MS/MS revealed that the application of trypsin@SiB in complex proteomic samples was also efficient. Reusability and long storage were also achieved by the new immobilized trypsin. All these demonstrated that this novel enzyme-immobilized reactor would be a promising strategy for high-speed digestion of proteomes and opened up a new avenue for applications of hollow silica bubbles in proteomics field.Conflict of interest
The authors declared no conflict of interest.Acknowledgements
This study was supported by the National Key Program for Basic Research of China (Grants 2012CB910603 and 2013CB911204), the National Key Program for Scientific Instrument and Equipment Development (Grants 2012YQ12004407, 2011YQ06008408 and 2013YQ14040506), and the National Natural Science Foundation of China (Grants 21275159 and 21235001).Notes and references
- J. Park, D. H. Cha, S. J. Lee, Y. N. Kim, Y. H. Kim and K. P. Kim, Exp. Mol. Med., 2011, 43, 427 CrossRef CAS PubMed.
- J. R. Whiteaker, C. W. Lin, J. Kennedy, L. M. Hou, M. Trute, I. Sokal, P. Yan, R. M. Schoenherr, L. Zhao, U. J. Voytovich, K. S. Kelly-Spratt, A. Krasnoselsky, P. R. Gafken, J. M. Hogan, L. A. Jones, P. Wang, L. Amon, L. A. Chodosh, P. S. Nelson, M. W. McIntosh, C. J. Kemp and A. G. Paulovich, Nat. Biotechnol., 2011, 29, 625 CrossRef CAS PubMed.
- B. R. Fonslow, B. D. Stein, K. J. Webb, T. Xu, J. Choi, S. K. Park and J. R. Yates, Nat. Methods, 2013, 10, 54 CrossRef CAS PubMed.
- G. W. Slysz, D. F. Lewis and D. C. Schriemer, J. Proteome Res., 2006, 5, 1959 CrossRef CAS PubMed.
- K. Venne, E. Bonneil, K. Eng and P. Thibault, Anal. Chem., 2005, 77, 2176 CrossRef CAS PubMed.
- K. Nakata, T. Ichibangase, R. Saitoh, M. Ishigai and K. Imai, Analyst, 2015, 140, 71 RSC.
- Q. Z. Luo, J. S. Page, K. Q. Tang and R. D. Smith, Anal. Chem., 2007, 79, 540 CrossRef CAS PubMed.
- J. R. Wiśniewski and M. Mann, Anal. Chem., 2012, 84, 2631 CrossRef PubMed.
- W. J. Qin, Z. F. Song, C. Fan, W. J. Zhang, Y. Cai, Y. J. Zhang and X. H. Qian, Anal. Chem., 2012, 84, 3138 CrossRef CAS PubMed.
- Z. Y. Hu, L. Zhao, H. Y. Zhang, Y. Zhang, R. A. Wu and H. F. Zou, J. Chromatogr. A, 2014, 1334, 55 CrossRef CAS PubMed.
- C. Fan, Z. M. Shi, Y. T. Pan, Z. F. Song, W. J. Zhang, X. Y. Zhao, F. Tian, B. Peng, W. J. Qin, Y. Cai and X. H. Qian, Anal. Chem., 2014, 86, 1452 CrossRef CAS PubMed.
- B. Jiang, K. G. Yang, Q. Zhao, Q. Wu, Z. Liang, L. H. Zhang, X. J. Peng and Y. K. Zhang, J. Chromatogr. A, 2012, 1254, 8 CrossRef CAS PubMed.
- S. Hermanová, M. Zarevúcká, D. Bouša, M. Pumera and Z. Sofer, Nanoscale, 2015, 7, 5852 RSC.
- X. J. Ren, H. H. Bai, Y. T. Pan, W. Tong, P. B. Qin, H. Yan, S. S. Deng, R. G. Zhong, W. J. Qin and X. H. Qian, Anal. Methods, 2014, 6, 2518 RSC.
- L. Chen, B. Wei, X. T. Zhang and C. Li, Small, 2013, 9, 2331 CrossRef CAS PubMed.
- X. Y. Mu, J. Qiao, L. Qi, P. Dong and H. M. Ma, ACS Appl. Mater. Interfaces, 2014, 6, 21346 CAS.
- Q. Wu, X. Wang, C. A. Liao, Q. C. Wei and Q. G. Wang, Nanoscale, 2015, 7, 16578 RSC.
- A. Butt, A. Farrukh, A. Ghaffar, H. Duran, Z. Oluz, H. ur Rehman, T. Hussain, R. Ahmad, A. Tahir and B. Yameen, RSC Adv., 2015, 5, 77682 RSC.
- H. M. Yuan, Y. Zhou, S. M. Xia, L. H. Zhang, X. D. Zhang, Q. Wu, Z. Liang and Y. K. Zhang, Anal. Chem., 2012, 84, 5124 CrossRef CAS PubMed.
- Q. Zhao, Y. Liang, H. M. Yuan, Z. G. Sui, Q. Wu, Z. Liang, L. H. Zhang and Y. K. Zhang, Anal. Chem., 2013, 85, 8507 CrossRef CAS PubMed.
- J. Tully, R. Yendluri and Y. Lvov, Biomacromolecules, 2016, 17, 615 CrossRef CAS PubMed.
- C. Chao, J. D. Liu, J. T. Wang, Y. W. Zhang, B. Zhang, Y. T. Zhang, X. Xiang and R. F. Chen, ACS Appl. Mater. Interfaces, 2013, 5, 10559 CAS.
- S. Nanayakkara, Z. Y. Zhao, A. F. Patti, L. Z. He and K. Saito, ACS Sustainable Chem. Eng., 2014, 2, 1947 CrossRef CAS.
- Z. Q. Chen, J. J. Zhang, S. Singh, P. Peltier-Pain, J. S. Thorson and B. J. Hinds, ACS Nano, 2014, 8, 8104 CrossRef CAS PubMed.
- Q. Yue, Y. Z. Li, M. Kong, J. C. Huang, X. J. Zhao, J. Liu and R. E. Williford, J. Mater. Chem., 2011, 21, 12041 RSC.
- M. Okamoto, H. Tsukada, S. Fukasawa and A. Sakajiri, J. Mater. Chem. A, 2015, 3, 11880 CAS.
- V. L. Schmit, R. Martoglio and K. T. Carron, Anal. Chem., 2012, 84, 4233 CrossRef CAS PubMed.
- V. L. Schmit, R. Martoglio, B. Scott, A. D. Strickland and K. T. Carron, J. Am. Chem. Soc., 2012, 134, 59 CrossRef CAS PubMed.
- J. J. Hu, R. N. Ma, F. Liu, Y. L. Chen and H. X. Ju, RSC Adv., 2014, 4, 28856 RSC.
- X. L. Liu, H. L. Liu, G. Y. Qing, S. T. Wang and X. M. Liang, J. Mater. Chem. B, 2014, 2, 2276 RSC.
- T. Blin, A. Kakinen, E. H. Pilkington, A. Lvask, F. Ding, J. F. Quinn, M. R. Whittaker, P. C. Ke and T. P. Davis, Polym. Chem., 2016, 7, 1931 RSC.
- Y. Gao, Y. Z. Wang, X. Xu, K. Ding and D. M. Yu, RSC Adv., 2014, 4, 63719 RSC.
- Y. Hu, N. N. Zhao, J. S. Li, W. T. Yang and F. J. Xu, J. Mater. Chem., 2012, 22, 21257 RSC.
- T. Pintauer and K. Matyjaszewski, Chem. Soc. Rev., 2008, 37, 1087 RSC.
- W. W. He, H. J. Jiang, L. F. Zhang, Z. P. Cheng and X. L. Zhu, Polym. Chem., 2013, 4, 2919 RSC.
- G. Moad, M. Chen, M. Häussler, A. Postma, E. Rizzardo and S. H. Thang, Polym. Chem., 2011, 2, 492 RSC.
- C. Guerrero-Sanchez, D. J. Keddie, S. Saubern and J. Chiefari, ACS Comb. Sci., 2012, 14, 389 CrossRef CAS PubMed.
- G. Huang, Z. Sun, H. Q. Qin, L. Zhao, Z. C. Xiong, X. J. Peng, J. J. Qu and H. F. Zou, Analyst, 2014, 139, 2199 RSC.
- Q. C. Cao, C. Ma, H. H. Bai, X. Y. Li, H. Yan, Y. Zhao, W. T. Ying and X. H. Qian, Analyst, 2014, 139, 603 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra12599a |
| This journal is © The Royal Society of Chemistry 2016 |