Mapping gold nanoparticles on and in edible leaves in situ using surface enhanced Raman spectroscopy†
Zhiyun Zhanga,
Huiyuan Guob,
Yingqing Dengb,
Baoshan Xingb and
Lili He*a
aDepartment of Food Science, University of Massachusetts, Amherst, MA, USA. E-mail: lilihe@foodsci.umass.edu; Fax: +1 413 545 1262; Tel: +1 413 545 5847
bStockbridge School of Agriculture, University of Massachusetts, Amherst, MA, USA
First published on 15th June 2016
Abstract
The increased prevalence of engineered nanomaterials (ENPs) in the environment and their potential toxicity require study on whether those engineered nanomaterials could possibly contaminate agricultural food and products. However, many techniques require invasive and complicated sample preparation procedures to detect and characterize engineered nanomaterials in complex matrices. Here, we present a non-destructive and label-free approach based on surface enhanced Raman spectroscopic (SERS) mapping technique to qualitatively detect and characterize gold nanoparticles (Au NPs), on and in spinach leaves in situ. We were able to detect the clearly enhanced signals from Au NPs at 15 to 125 nm on and in spinach leaves. Peak characterizations revealed the aggregation status of Au NPs and their interactions with plant biomolecules, such as chlorophylls and carotenoids. This developed approach will open a new analytical platform for various researches on studying ENPs' adhesion and accumulation.
Introduction
In recent years, engineered nanoparticles, such as silver and copper, are increasingly used in agriculture due to their antimicrobial properties. For example, silver nanoparticles (Ag NPs) have been widely used to protect crops against plant pathogens and pests.1 As of 2010, there had been more than 110 officially registered Ag NPs containing pesticides used for agricultural, environmental, medical, and home purposes in the US.2 Copper based pesticides, including copper nanoparticles (Cu NPs) have also been used widely as fungicides in vineyards and farms.1 However, the use of these NPs in agriculture may pose some potential risks. A number of studies show that certain engineered nanoparticles (ENPs) are more toxic to microbes, plants, animal and/or human cells compared to their ionic or bulk counterparts.3 The increasing prevalence of ENPs within agriculture and food products and their potential toxicity have urged researchers to study how those ENPs could possibly contaminate the environment and bioaccumulate along the food chain, and to evaluate their chemical and biological effect on human health and the environment. However, research on ENPs as emerging contaminants is still a new field.4,5 Some studies suggested that NPs can accumulate in plants after foliar exposure6–8 and may be able to translocate from soil to plant tissues.9–11 The interactions between NPs and plants depend on size and surface charge of NPs,12,13 and are also plant species-specific.14,15 These bioaccumulated NPs can enter into food chains; and can then be transferred to consumers, causing unknown risks to sensitive receptors.5Various techniques have been used for detection and characterization of ENPs in planta, such as inductively coupled plasma based methods,6–8,14 X-ray absorption spectroscopy,13,15–17 and electron microscopy.6–8 However, the majority of these techniques require complex digestion and extraction procedures for analyzing NPs from complex samples.18 Synchrotron X-ray fluorescence microscopy has been used for in situ mapping and speciation of CeO2 in kidney beans19 and cucumber roots.20 However, there are certain disadvantages of this technique, including the additional absorption of characteristic X-rays by the sample itself on their path to the detector system, especially for low energy X-rays or where samples are particularly dense or large (exceeding a few hundred micrometers), the absorption effects can be severely influenced.21 In addition, access to synchrotron facilities is limited. Thus, the development of a rapid and reliable method for the detection and characterization of ENPs in complex matrices is needed.
Surface enhanced Raman spectroscopy (SERS) is a combined technique that involves both Raman spectroscopy and nanotechnology. Noble metals, such as NP Au, Ag, and Cu, can significantly enhance the Raman signals of the molecules that are in close vicinity of metal surfaces. This is because the excitation of localized surface plasmon resonance on noble metal NPs can generate a large electromagnetic field that increases the Raman cross section from molecules that are in close proximity (∼10 nm) of a noble metal nanostructure.22 Due to its improved sensitivity, SERS has been applied for the detection of various chemical and biological targets in many areas, such as medical diagnosis,23 food24,25 and environmental safety.26 In addition, SERS mapping has been applied as an imaging tool for intracellular studies. For example, Rodríguez-Lorenzo et al. utilized SERS-encoded gold nanostars for intracellular mapping.27 Ando et al. reported a dynamic SERS imaging method based on Au NPs being applied to study dynamic biological functions in living cells, such as membrane protein diffusion, nuclear entry, and rearrangement of cellular cytoskeleton.28 Shen et al. also found that SERS can be used as a rapid and non-invasive imaging technique to monitor the distribution of 4-mercapto benzoic acid tagged carbon-encapsulated Au–Ag NPs inside the leaf.29 To date, however, most of the analytical targets for SERS are chemical and biological compounds.
Herein, we aimed at NPs rather than the chemical and biological targets. The objective is to evaluate the SERS technique for in situ, non-destructive and label-free detection of Au NPs on and in spinach leaves after foliar exposure and characterization of the interaction between Au NPs and spinach. The innovation of this study lies in the use of intrinsic enhanced SERS signals from the biomolecules to detect the presence of noble metallic nano-contaminants and determine their final fate in plants. Coupled with mapping technique, this SERS method can spatially image the heterogeneous distribution of NPs on and in spinach leaves in situ and non-destructively. Au NPs were chosen as the model NPs to evaluate and demonstrate method feasibility, because they can easily be synthesized with a uniform size and shape, have low environmental background level, and are chemically inert and stable in size or shape under various environmental and biological conditions.14 Spinach was selected as the model plant because of its large consumption worldwide and large shoot surface area, which is ideal for foliar study.
Experimental section
Materials
Gold(III) chloride trihydrate and hydroquinone were purchased from Sigma-Aldrich (St. Louis, MO). Sodium citrate dehydrate was purchased from Fisher Scientific (Pittsburgh, PA). Organic spinach leaves were purchased from a local grocery store in Amherst, MA and transferred to the Chenoweth Lab at University of Massachusetts Amherst. All spinach leaves were stored at 4 °C and used within 1 day. All leaves were washed with deionized water (Barnstead MicroPure system, Fisher Scientific Co., Pennsylvania) with a pH of 6.Fabrication and characterization of Au NPs
15 nm Au NPs were synthesized by the Turkevich method and 35 to 125 nm Au NPs were synthesized by the hydroquinone reduction and seed-mediated growth method.30Transmission electron microscopy (TEM, JEOL JEM-2000FX) was used to characterize the synthesized Au NPs. In order to completely disperse the Au NPs, we used probe sonicated (Branson 2800) for our Au NPs with 15 minutes before dropping on the copper grids. The sizes of synthesized Au NPs (n = 100) were measured using the ImageJ Software (NIH, Bethesda, MD) based on acquired TEM images (Fig. S1†). We also measured the particle size distributions of the Au NPs samples using a dynamic light scattering instrument (Mastersizer 2000, Malvern Instruments). The surface charge of Au NPs was determined by using a particle electrophoresis instrument (Zetasizer Nano ZS series, Malvern Instruments) (Table S1†). UV-vis absorption spectra of the Au NPs samples were recorded on a SpectraMax spectrophotometer (Molecular Devices, LLC., CA) in the range 350–750 nm with 10 nm resolution. Plastic cuvettes with a 1 cm optical length were used (Table S1†).
Preparation for in situ study of Au NPs adsorbed on spinach leaf surfaces
To study the Au NPs adsorbed on spinach leaf surfaces, 3 mL Au NPs of different concentrations (50 and 5 mg L−1) and sizes (15, 35, 80, and 125 nm) were prepared in Petri-dishes. The concentrations of the Au NPs (50 and 5 mg L−1) used in this study are based on the concentrations of Ag NPs currently used in the commercial pesticide products in the US market. Then, fresh spinach leaves were immersed into these solutions and incubated for 30 minutes on the Fisher Scientific™ Nutating Mixers (Fisher Scientific Co., PA) at the low speed of 24 rpm to ensure the leaves fully exposure to Au NPs. After that, the leaves were gently rinsed with deionized water for 3 minutes and air-dried in the hood under room temperature. Spinach leaves without Au NPs were used as a control. Bright field light scattering images, Raman images, and representative Raman spectra were then collected.Preparation for in situ study of Au NPs penetrated into spinach leaves
To study the penetration of Au NPs into spinach leaves, 10 μL Au NPs (50, 200 mg L−1) were dropwise deposited on the leaf surfaces in predetermined areas. The spinach leaves that were treated with Au NPs were air-dried in the hood at room temperature. Raman images, and representative Raman spectra were collected immediately.Raman instrumentation and data analysis
A DXR Raman microscope (Thermo Fisher Scientific, Madison, WI) with a 780 nm laser and 10×, 20× confocal microscope objectives were used in this study. Each spectrum was scanned from 3400 to 400 cm−1 with 5 mW laser power and 2 s exposure time. Raman maps were integrated based on the characteristic peaks in the Raman spectra using the atlμs function in the OMINCS software (Thermo Fisher Scientific). For the surface study, Raman mapping was applied with a 50 μm slit aperture to maximize the signals. To compare Raman and optical images, the step size is 10 μm step size and each image contains 100 spots. To map Au NPs of different sizes (15–125 nm), the step size is 40 μm and each image contains 360 spots. In this way, we can quickly scan the representative area within 30 min. For the penetration study, Raman mapping was applied with a 50 μm pinhole aperture to control the confocal depths. The step size in X direction is 10 μm and in Z direction is 10 μm, and each image contains 150 spots. The instrumentation parameters (laser power and exposure time) were optimized to achieve sensitive and rapid detection without damaging the leaf tissues.Transmission electron microscopy characterization of Au NPs in spinach
Au NPs distribution in spinach leaves was observed by TEM (JEOL, JEM-2200FX). Spinach leaves were prepared by fixation, dehydration, infiltration and polymerized at 60 °C for 24 hours.31 The ultrathin sections (90 nm) were cut and placed on the grid. Finally, TEM (200 kV) was used to observe the specimens.Results and discussion
In situ detection and characterization of Au NPs on spinach leaves
Fig. 1a and c show bright field light scattering images of spinach leaves without and with Au NPs (35 nm, 50 mg L−1). As shown in Fig. 1c, Au NPs were heterogeneously distributed on the spinach leaves' surfaces. This uneven distribution of Au NPs is likely due to the complex structures of the spinach leaves' surface. The bright color of Au NPs is a result of their surface plasmon (SPR).32Fig. 1b and d are the corresponding Raman images which were constructed based on the highest peak at ∼1525 cm−1. The peak assignments for the normal Raman spectra of carotenoids and plant leaves have been previously reported.33,34 Three major peaks (1525, 1156 and 1005 cm−1) have been identified as carotenoids, which are presented in the plant leaves. Among these three peaks, the 1525 cm−1 is the highest. We also extracted all the pigments (chlorophylls and carotenoids) and measured their SERS signals. Our results (Fig. S3†) agreed with the references. Therefore, the 1525 cm−1 peak is most likely from carotenoids. As in other imaging techniques, it is critically important to identify and subtract background signals to minimize matrix interference. Here we set 200 counts (at 1525 cm−1) as the baseline for background subtraction for the best results with 2% false positive and 5% false negative (Fig. S2†). As shown in Fig. 1d, when Au NPs were on the leaves, spots with higher intensity were clearly shown in different colors other than blue, which indicates the presences of Au NPs on leaf surfaces. These Au NPs are mainly Au NPs aggregates, as individual Au NPs have very weak enhancement.35 The spectra varied from spot to spot with different patterns and intensities, indicating that both the distribution and local environment of Au NPs were quite heterogeneous. In addition, the non-flat surface would also result in the orientation difference between the laser and Au NPs, which would contribute to the spectral variance. The assignment of SERS peaks is very difficult compared with normal Raman, as molecules can interact with NPs in many different ways. Generally speaking, only the molecules adsorbed (interacted) on the Au NPs were most enhanced. This is because the enhancement is highly distance dependent. Other molecules may be surrounding the Au NPs; however, if the distance is larger than ∼10 nm, there is no enhancement at all.36 The selected SERS spectra (Fig. 1f and g) show enhanced peaks that are similar to the normal Raman spectra (Fig. 1e), which indicates the interactions between the Au NPs and leaf chlorophylls and carotenoids. To verify this, we extracted chlorophylls and carotenoids from spinach leaves and mixed them with Au NPs. The resulting SERS spectra (Fig. S3†) show similar characteristic peaks to the in situ spectra (Fig. 1f and g), demonstrating the interaction between Au NPs and plant pigments in situ. In the literature review, we found two possible mechanisms for the interaction between Au NPs and chlorophylls. One study indicates the negatively charged Au NPs are bound to the magnesium metal center of chlorophylls, which is coordinatively unsaturated.37 Another study demonstrates the formation of Au NPs and chlorophylls complex is due to the ligand-exchange reaction between Au NPs and nitrogens of chlorophylls via nonbonding electrons.38 The binding constant for Au NPs and chlorophyll is very high, ∼105 M−1 and the amount of chlorophylls in spinach leaves is about 1–2%.39 Therefore, chlorophylls are highly likely to interact with Au NPs and thus be reflected in the SERS spectra. Other peaks have also been observed, which indicates the complexity of the biomolecules co-adsorbed or close to the Au NPs. For example, some carotenoids peaks were observed in the in situ spectra as well. In addition, a peak at 2130 cm−1 was observed after the application of Au NPs on spinach leaves. Since no peak at 2130 cm−1 was observed on spinach leaves without Au NPs, we assume this peak may come from Au NPs, which may prove that the spectra we obtained are involved with Au NPs. The origin of this peak is unknown. In addition, some spots with high intensity (e.g. Fig. 1h) were also observed. The SERS spectrum of hot spot was significantly different than the others, with broader shifts at around 1000–1700 cm−1. This may be due to significant aggregation of NPs that produced stronger and multiple localized SPR which enhanced and broadened the carbon peaks and thus reduced the characteristic information. We observed a similar phenomenon when measuring Au NPs aggregates on a gold coated glass slide (Fig. S4†). Although those super-hot spots can be used to determine both the presence and aggregation state of Au NPs, the characteristic information of the NPs–leaf interactions may not be reflected.
Given the limitation of all the micro-imaging techniques, we could only look at a small area under the Raman microscope. Therefore, it is important to select an area that is statistically representative of the entire target. Though we can scan at a very fine step over the entire leaves to collect all the information, it is too time-consuming, impractical, and not statistically meaningful. Comparing the Raman image with the optical image, we found most parts of these two images matched fairly well. Most of the Au NPs shown in the optical image also produced signals in similar positions in the Raman image, though some spots were missed because the set step size (10 μm) is larger than the laser spot (3 μm). The intensity of the Au NPs in these two images did not correlate. The intensity of Au NPs in the optical image is mainly based on the amount, while the intensity of SERS signals also depends on some different factors, such as amount, aggregation, and interactions. In addition, since the surfaces of spinach leaves are not flat, in a scanning area, some parts of the area may be in focus and some may be out of focus. Thus, if the part of the scanning areas is out of focus, even with a large amount of Au NPs, the SERS signals may also be weak. In addition, some undetected NPs not shown in the optical image were detected by using SERS, probably due to the penetration ability and increased sensitivity of the laser. Compared with our previous study that used a Raman reporter (ferbam) as the indicator to detect and quantify Ag NPs in liquid and semi-liquid matrix,40 no indicator was used in this study. This is because the purpose of this study is not only to detect the Au NPs, but also to investigate whether we are able to characterize the interactions between the Au NPs and plant biomolecules based on the SERS signals. If an indicator was used, the sensitivity and quantitative ability of detection may be improved; however, we lose the information about plant–NPs interactions.
Raman mapping of Au NPs of various sizes on spinach leaves
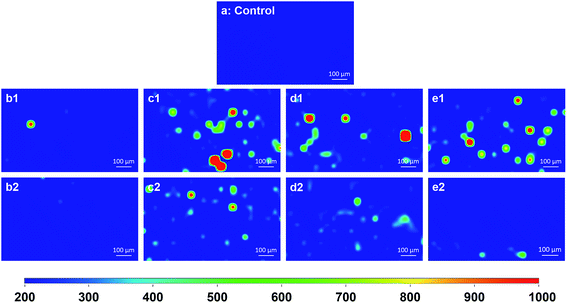
To evaluate the mapping method for measuring Au NPs of different sizes (15–125 nm) on spinach leaves, we randomly picked an area on the leaves with the size of 920 μm × 560 μm and used a relative large step size (40 μm), which resulted in 360 spots per image. In this way, we were able to quickly scan the representative area within 30 min.In Fig. 2b1–e1, after being contaminated with Au NPs at 50 mg L−1, it is interesting to find that, except 15 nm Au NPs treated group, strong SERS signals were obtained from each of the other three groups, which indicates the presence of Au NPs on these spinach leaves. The intensity of SERS signals is strongly determined by the following aspects: (1) the aggregation status (hot spots) of Au NPs; (2) the size of Au NPs in the aggregation; and (3) the number of NPs in the probed area. In this study, we deposited different sizes of Au NPs under the same mass, which means the number of Au NPs with smaller size is larger than those with bigger size. As shown in Table S1,† 15 nm NPs have the lowest SPR, therefore, they have the least enhancement factor even in the aggregation status. Although their number is the largest, it is still very challenging to detect them. Furthermore, when we decreased the concentration of Au NPs to 5 mg L−1 (Fig. 2b2–e2), although SERS intensity became weaker, 35, 80 and 125 nm Au NPs were still detectable in situ. This data demonstrated that we were able to map various sizes of Au NPs on spinach leaves in situ. Although increasing exposure time and/or laser power may enhance the sensitivity, it may cause potential damage to the leaves and significantly increase the time for image analysis using this mapping technique.
In situ detection and characterization of Au NPs in spinach leaves
There are two non-destructive approaches of using the confocal Raman spectroscopy to detect and characterize Au NPs in spinach leaves in situ. The first approach is to scan the area maps (XY) at different depth. Fig. 3a–c show the Raman images of three different depths (0, 10, and 20 μm). Hot spots with strong signals in 10 μm and 20 μm depth image were clearly observed, indicating that Au NPs were able to penetrate into the spinach leaves. Compared with 0 and 10 μm images, the number of spots with high intensity significantly decreased at 20 μm depth, which means there are decreasing amounts and less aggregation of Au NPs in deeper areas. Looking into the selected SERS spectra at different depths, the 0 and 10 μm spectra (Fig. 3d and e) do not have characteristic peaks but a broad peak between 800–1600 cm−1, which indicates the NP–NP interaction (aggregation), while the spectrum of Fig. 3f shows clear enhanced peaks of carotenoids and chlorophylls, which demonstrates NP–pigment interactions. We also characterized other spots in the 20 μm depth images and found most of them showing various patterns combining the characteristic peaks of carotenoids and chlorophylls (Fig. S5†). This indicates strong interactions between Au NPs and plant pigments.The second approach is to scan the area map vertically (XZ) to get more direct information on the penetration depth of Au NPs. Based on the previous report, it was estimated that the thickness of a spinach leaf was normally from 300 to 600 μm.41 Thus, we scanned from the top to 300 μm in depth to study the penetration depth of 35 nm Au NPs. Multiple images were collected randomly on the leaf surfaces, and three representative images were shown in Fig. 3h–j. Compared to the control (without Au NPs), these images show enhanced signals though varied with penetration depth from 80–150 μm. The variation of the penetration depth may be caused by spatially heterogeneous leaf structures and properties, including spinach leaves' wax coverage, surface wettability, stomata geometry and permeability, and so on.42,43 Several studies demonstrated that stomatal43 and cuticular pathways7,8 may enable ENPs accumulation in plant leaves through foliar exposure. In this study, we observed both pathways for Au NPs penetrating into spinach leaves as shown in Fig. S6.† In terms of penetration depth, there is no significant difference between these two pathways. But stomata may allow more Au NPs to penetrate in some cases, as indicated by intense signals observed in the depth image.
We also did a validation study by cutting the leaf and scanning the cross-sections. As shown in Fig. S7,† the strongest SERS intensity was observed mainly at around 30 μm depth. With the depth increasing, although the Raman intensity in each layer became weaker, up to 240 μm, the intensity of Raman spectrum was still around 400 counts. The depth profile obtained from this method is deeper than the previous method. One possibility is the under-estimation of the confocal scanning, which resulted from decreased penetration ability of laser at further depth and heterogeneous structure of spinach leaves.44 However, the result from the cutting method may be over-estimated as the pressure of cutting may artificially enhance the Au NPs' accumulation. Nevertheless, it may not be practically meaningful to estimate the absolute penetration depth. These results demonstrate that the Raman mapping technique can be used to measure Au NPs in spinach leaves in situ.
Raman mapping of Au NPs of various sizes in spinach leaves
We then applied the vertical mapping approach to study the size effect on NP penetration. Four sizes (15, 35, 80, and 125 nm) and two concentrations (50 and 200 mg L−1) were used. For each size and concentration, at least five mappings were collected below the cuticle. The deepest penetration depth images were shown in Fig. 4. Au NPs of all sizes can penetrate into spinach leaves to different depths. In addition, we observed a size dependent penetration effect. The 125 nm Au NPs were found remaining mostly close to the surface, while the 80 and 35 nm Au NPs penetrated into approximately 100 and 150 μm in depth, respectively. This is probably due to the diffusion coefficients, which are inversely proportional to the radius of the permeant.43,45 Thus, it is reasonable to hypothesize that the part of Au NPs that penetrated into deep area might come from the Au NPs with smaller size. The reason for the low penetration depth of 15 nm observed in the image is likely due to the weaker signals from 15 nm which made it difficult to track these Au NPs in deeper depth, although they may penetrate the deepest. In addition to the size effect, we observed the Au NPs at higher concentrations penetrated deeper than lower concentrations and the signal intensities were higher than those of low concentrations in the Raman images. One study also found a similar effect of concentration on the penetration depth.46 However, this may also be influenced by the sensitivity of the SERS mapping techniques which captured more signals when the concentration was higher.Validation of the SERS mapping using TEM-EDS
To validate the SERS mapping results, TEM-EDS was used to observe and confirm Au NPs in spinach leaves. Fig. 5a and b show TEM images in a vertical section of a spinach leaf treated with 35 nm Au NPs (200 mg L−1). It was found that Au NPs penetrated into the spinach leaf interior and were distributed both outside and inside of the leaf cell walls. Furthermore, a considerable amount of NPs was distributed in and around the chloroplasts, structures that contain mainly chlorophylls and carotenoids. This may further confirm the strong signals from plant pigments observed in the previous Raman spectra. In addition, many Au NPs were shown in aggregated status inside the leaf tissues, which also confirms the observation from previous Raman spectra. Other studies also reported that certain ENPs can attach to the surface of chloroplasts47 and even enter chloroplasts.48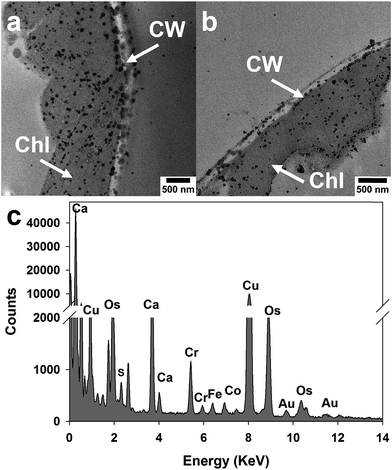 | ||
| Fig. 5 TEM-EDS images of spinach leaves treated with 35 nm Au NPs (200 mg L−1). Chloroplast (Chl) and Cell wall (CW). | ||
Conclusion
In this current work, we developed an innovative, simple, and rapid approach using SERS mapping technique to detect and characterize different sizes of Au NPs on and in spinach leaves in situ. The detection was based on the hot spots produced by Au NPs on and in spinach leaves which can be clearly captured using Raman mapping without any sample preparation steps. The intensity and spectral pattern of hot spots reveal NP aggregation status as well as the interactions between Au NPs and plants. The Raman intensity of characteristic peaks from chlorophylls and carotenoids were enhanced, which indicates the interactions between Au NPs and these plant bio-components. TEM-EDS also verified the interaction between Au NPs and chloroplast. To the best of our knowledge, it is the first study that explored and applied SERS mapping for detection and characterization of NP contaminants attaching onto and internalizing into fresh produce. We foresee this effective and transformative technique to open a new and exciting analytical window for rapidly detecting the presence of ENPs (especially Au, Ag, and Cu) in complex biological samples (such as plant leaves, biofilm, human and animal skins, etc.). More importantly, the interactions of ENPs with bio-components in situ can be investigated, which will greatly facilitate the understanding of ENPs' adhesive and uptake mechanisms, and further promote the understanding the behavior and fate of ENPs. We will further explore and apply this method to future studies of other ENPs (e.g., Ag and Cu NPs) and their interactions with plant tissues.Conflict of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the National Institute of Food and Agriculture of the U.S. Department of Agriculture (USDA-NIFA, grant no.: 2015-67017-23070).References
- A. Gogos, K. Knauer and T. D. Bucheli, J. Agric. Food Chem., 2012, 60, 9781–9792 CrossRef CAS PubMed.
- L. L. Bergeson, Environ. Qual. Manag., 2010, 19, 73–82 CrossRef.
- D. M. Whitacre and F. A. Gunther, Reviews of environmental contamination and toxicology, Springer, New York, 2012 Search PubMed.
- C. Remédios, F. Rosário and V. Bastos, J. Bot., 2012, 2012, 1–8 CrossRef.
- C. M. Rico, S. Majumdar, M. Duarte-Gardea, J. R. Peralta-Videa and J. L. Gardea-Torresdey, J. Agric. Food Chem., 2011, 59, 3485–3498 CrossRef CAS PubMed.
- J. Hong, J. R. Peralta-Videa, C. Rico, S. Sahi, M. N. Viveros, J. Bartonjo, L. Zhao and J. L. Gardea-Torresdey, Environ. Sci. Technol., 2014, 48, 4376–4385 CrossRef CAS PubMed.
- C. Larue, H. Castillo-Michel, S. Sobanska, L. Cécillon, S. Bureau, V. Barthès, L. Ouerdane, M. Carrière and G. Sarret, J. Hazard. Mater., 2014, 264, 98–106 CrossRef CAS PubMed.
- C. Larue, H. Castillo-Michel, S. Sobanska, N. Trcera, S. Sorieul, L. Cécillon, L. Ouerdane, S. Legros and G. Sarret, J. Hazard. Mater., 2014, 273, 17–26 CrossRef CAS PubMed.
- J. L. Gardea-Torresdey, C. M. Rico and J. C. White, Environ. Sci. Technol., 2014, 48, 2526–2540 CrossRef CAS PubMed.
- A. D. Servin, M. I. Morales, H. Castillo-Michel, J. A. Hernandez-Viezcas, B. Munoz, L. Zhao, J. E. Nunez, J. R. Peralta-Videa and J. L. Gardea-Torresdey, Environ. Sci. Technol., 2013, 47, 11592–11598 CrossRef CAS PubMed.
- T. K. Darlington, A. M. Neigh, M. T. Spencer, O. T. Nguyen and S. J. Oldenburg, Environ. Toxicol. Chem., 2009, 28, 1191–1199 CrossRef CAS PubMed.
- J. S. Bozich, S. E. Lohse, M. D. Torelli, C. J. Murphy, R. J. Hamers and R. D. Klaper, Environ. Sci.: Nano, 2014, 1, 260–270 RSC.
- J. D. Judy, J. M. Unrine, W. Rao, S. Wirick and P. M. Bertsch, Environ. Sci. Technol., 2012, 46, 8467–8474 CrossRef CAS PubMed.
- Z.-J. Zhu, H. Wang, B. Yan, H. Zheng, Y. Jiang, O. R. Miranda, V. M. Rotello, B. Xing and R. W. Vachet, Environ. Sci. Technol., 2012, 46, 12391–12398 CrossRef CAS PubMed.
- M. L. López-Moreno, G. De La Rosa, J. A. Hernández-Viezcas, J. R. Peralta-Videa and J. L. Gardea-Torresdey, J. Agric. Food Chem., 2010, 58, 3689–3693 CrossRef PubMed.
- G. Uzu, S. Sobanska, G. Sarret, M. Muñoz and C. Dumat, Environ. Sci. Technol., 2010, 44, 1036–1042 CrossRef CAS PubMed.
- J. Kurepa, T. Paunesku, S. Vogt, H. Arora, B. M. Rabatic, J. Lu, M. B. Wanzer, G. E. Woloschak and J. A. Smalle, Nano Lett., 2010, 10, 2296–2302 CrossRef CAS PubMed.
- K. Tiede, A. B. A Boxall, S. P. Tear, J. Lewis, H. David and M. Hassellov, Food Addit. Contam., Part A, 2008, 25, 795–821 CrossRef CAS PubMed.
- S. Majumdar, J. R. Peralta-Videa, S. Bandyopadhyay, H. Castillo-Michel, J. A. Hernandez-Viezcas, S. Sahi and J. L. Gardea-Torresdey, J. Hazard. Mater., 2014, 278, 279–287 CrossRef CAS PubMed.
- X. Gui, X. He, Y. Ma, P. Zhang, Y. Li, Y. Ding, K. Yang, H. Li, Y. Rui, Z. Chai, Y. Zhao and Z. Zhang, RSC Adv., 2015, 5, 4554–4560 RSC.
- T. Punshon, M. Lou Guerinot and A. Lanzirotti, Ann. Bot., 2009, 103, 665–672 CrossRef CAS PubMed.
- C. L. Haynes, A. D. McFarland and R. P. Van Duyne, Anal. Chem., 2005, 77, 338A–346A CrossRef CAS.
- T. Vo-Dinh, F. Yan and M. B. Wabuyele, J. Raman Spectrosc., 2005, 36, 640–647 CrossRef CAS.
- J. Zheng and L. He, Compr. Rev. Food Sci. Food Saf., 2014, 13, 317–328 CrossRef CAS.
- A. P. Craig, A. S. Franca and J. Irudayaraj, Annu. Rev. Food Sci. Technol., 2013, 4, 369–380 CrossRef CAS PubMed.
- R. A. Halvorson and P. J. Vikesland, Environ. Sci. Technol., 2010, 44, 7749–7755 CrossRef CAS PubMed.
- L. Rodríguez-Lorenzo, Z. Krpetic, S. Barbosa, R. A. Alvarez-Puebla, L. M. Liz-Marzán, I. A. Prior and M. Brust, Integr. Biol., 2011, 3, 922–926 RSC.
- J. Ando, K. Fujita, N. I. Smith and S. Kawata, Nano Lett., 2011, 11, 5344–5348 CrossRef CAS PubMed.
- A. Shen, J. Guo, W. Xie, M. Sun, R. Richards and J. Hu, J. Raman Spectrosc., 2011, 42, 879–884 CrossRef CAS.
- C. D. Walkey, J. B. Olsen, H. Guo, A. Emili and W. C. W. Chan, J. Am. Chem. Soc., 2012, 134, 2139–2147 CrossRef CAS PubMed.
- J. J. Bozzola and L. D. Russell, Electron microscopy: principles and techniques for biologists, Jones & Bartlett Learning, Burlington, 1999 Search PubMed.
- S. Eustis and M. A. El-Sayed, Chem. Soc. Rev., 2006, 35, 209–217 RSC.
- H. Schulz and M. Baranska, Vib. Spectrosc., 2007, 43, 13–25 CrossRef CAS.
- R. Withnall, B. Z. Chowdhry, J. Silver, H. G. M. Edwards and L. F. C. de Oliveira, Spectrochim. Acta, Part A, 2003, 59, 2207–2212 CrossRef.
- A. M. Schwartzberg, C. D. Grant, A. Wolcott, C. E. Talley, T. R. Huser, R. Bogomolni and J. Z. Zhang, J. Phys. Chem. B, 2004, 108, 19191–19197 CrossRef CAS.
- S. Laing, K. Gracie and K. Faulds, Chem. Soc. Rev., 2016, 45, 1901–1918 RSC.
- S. Barazzouk, P. V. Kamat and S. Hotchandani, J. Phys. Chem. B, 2005, 109, 716–723 CrossRef CAS PubMed.
- S. Barazzouk, L. Bekalé and S. Hotchandani, J. Mater. Chem., 2012, 22, 25316–25324 RSC.
- U. Kidmose, M. Edelenbos, L. P. Christensen and E. Hegelund, J. Chromatogr. Sci., 2005, 43, 466–472 CAS.
- H. Guo, Z. Zhang, B. Xing, A. Mukherjee, C. Musante, J. C. White and L. He, Environ. Sci. Technol., 2015, 49, 4317–4324 CrossRef CAS PubMed.
- V. Amiard, K. E. Mueh, B. Demmig-Adams, V. Ebbert, R. Turgeon and W. W. Adams, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 12968–12973 CrossRef CAS PubMed.
- T. Eichert and J. Burkhardt, J. Exp. Bot., 2001, 52, 771–781 CrossRef CAS PubMed.
- T. Eichert, A. Kurtz, U. Steiner and H. E. Goldbach, Physiol. Plant., 2008, 134, 151–160 CrossRef CAS PubMed.
- K. C. Gordon and C. M. McGoverin, Int. J. Pharm., 2011, 417, 151–162 CrossRef CAS PubMed.
- Z. Zhang, F. Kong, B. Vardhanabhuti, A. Mustapha and M. Lin, J. Agric. Food Chem., 2012, 60, 10762–10767 CrossRef CAS PubMed.
- V. Fernández and T. Eichert, CRC Crit. Rev. Plant Sci., 2009, 28, 36–68 CrossRef.
- L. Van Nhan, C. Ma, Y. Rui, S. Liu, X. Li, B. Xing and L. Liu, Sci. Rep., 2015, 5, 11618 CrossRef PubMed.
- L. Zheng, F. Hong, S. Lu and C. Liu, Biol. Trace Elem. Res., 2005, 104, 83–92 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra11748a |
| This journal is © The Royal Society of Chemistry 2016 |