Rare sugar production by coupling of NADH oxidase and L-arabinitol dehydrogenase†
Hui Gaoab,
Tae-Su Kima,
Primata Mardinaa,
Pengji Zhoub,
Fei Wen*b and
Jung-Kul Lee*a
aDepartment of Chemical Engineering, Konkuk University, Seoul 05029, Republic of Korea. E-mail: jkrhee@konkuk.ac.kr; Fax: +82-2-458-3504; Tel: +82-2-450-3505
bDepartment of Chemical Engineering, University of Michigan, Ann Arbor, MI 48109, USA. E-mail: feiwenum@umich.edu; Fax: +1-734-763-0459; Tel: +1-734-764-8723
First published on 4th July 2016
Abstract
An efficient biocatalytic cell-free system containing L-arabinitol dehydrogenase (LAD) for L-arabinitol oxidation and NADH oxidase (Nox) for cofactor regeneration was successfully constructed and used for L-rare sugar production. The recombinant LAD (HjLAD) from Hypocrea jecorina suffered from NADH inhibition. Applying Nox to the cell-free HjLAD system drove the thermodynamically unfavorable equilibrium from L-arabinitol to L-xylulose, along with the regeneration of the oxidized cofactor NAD+. Response surface methodology was used to optimize the conditions for L-xylulose production. The optimal conditions for biocatalytic production of L-xylulose were found to be 4.9 U mL−1 HjLAD, 8.2 mM NAD+ at pH 8.0, and 30.9 °C; a high conversion rate of 78.4% was achieved under these conditions. We demonstrate a cell-free enzyme coupling system enabling efficient cofactor recycling and providing high yields of L-xylulose. The results indicate that this coupling system provides a new biocatalytic method for L-rare sugar production.
1. Introduction
Rare sugars and their derivatives, specifically monosaccharides, rarely occur in nature. They have various applications in the pharmaceutical, nutrition, and food industries as nucleoside analogs, precursors, no-calorie sweeteners, and bulk agents, despite their low natural abundance.1,2 Nucleoside analogs of L-sugars are used as antiviral drugs in the treatment of hepatitis B and HIV, and they also show promise as anticancer and cardioprotective agents.3–5 L-Xylulose, one of the rare sugars, is a ketopentose that exists in very low concentrations in nature. It is an intermediate in many prokaryotic and eukaryotic metabolic pathways6–8 and has been used as an inhibitor of glycosidases and as a reliable marker for acute or chronic hepatitis or liver cirrhosis.5,9 Therefore, it is important to synthesize sufficient amounts of L-xylulose efficiently and economically to meet its application needs.NAD(P)+-dependent polyol dehydrogenases are important catalysts that have revolutionized the conversion and utilization of biorenewable feedstocks for the production of value-added chemicals, fuels, and pharmaceutical intermediates.5,10 L-Sugars can be produced using polyol NAD(P)+-dehydrogenases, but a major challenge for the practical application is the need for a stoichiometric amount of the expensive nicotinamide cofactor NAD(P)H or NAD(P)+ involved in enzyme-catalyzed oxidoreductions. Cofactors are mediators carrying electrons and are consumed stoichiometrically during the redox reaction. Owing to the relatively high cost of NAD(P)H or NAD(P)+, they have to be regenerated and reused to reduce the overall production cost. Cofactor regeneration is an important solution to the problem of implementing complex cofactor requiring enzymatic reactions at the industrial scale. A number of chemical, electrochemical, photochemical, and enzymatic methods for regenerating the nicotinamide cofactors NAD(P)H or NAD(P)+ have been established and recently reviewed.11–14 Among them, the enzymatic methods seem to be the most convenient and useful.11
Biological production of L-xylulose from xylitol has been established using resting cells of bacteria such as Pantoea ananatis,15,16 Alcaligenes sp. 701B,17 and Bacillus pallidus.5,9 Cloning of xylitol 4-dehydrogenase (XDH) genes from P. ananatis and B. pallidus was also reported and the recombinant strains were used to produce L-xylulose. However, the thermodynamic equilibrium of XDH between xylulose and xylitol has been shown to be strongly on the side of xylitol.15 Over 100 mM xylitol present in the reaction will inhibit the reaction toward L-xylulose production.15 Previous reports showed that the highest L-xylulose yield from xylitol was 85% at an initial low substrate concentration of 2% (∼131 mM) at 50 °C.9 The low L-xylulose yields from xylitol 4-dehydrogenase catalyzed reactions may also be due to NADH inhibition, which was indicated in xylitol 4-dehydrogenase kinetic mechanism studies.18–20 Nidetzky et al. performed a detailed steady-state kinetic analysis of xylitol dehydrogenase using sorbitol as substrate and found that dissociation of NADH appears to be the main rate-limiting step under substrate-saturated reaction conditions.20 NADH showed a competitive inhibition pattern at both non-saturating and saturating concentrations of substrate with a Ki value of 10 μM.20 Polyol dehydrogenase, like D-sorbitol dehydrogenase,19,21 L-glutamate dehydrogenase,22 and mannitol dehydrogenase,23,24 may share the property of NADH inhibition. However, this property has not been described as a limitation factor in the application of polyol dehydrogenase as a biocatalyst for L-xylulose production.
In the present study, we demonstrate a simple, highly efficient, and economical cell-free system comprising L-arabinitol dehydrogenase from Hypocrea jecorina (HjLAD) coupled with an NADH oxidase from Streptococcus pyogenes (SpNox) for regeneration of the cofactor NAD+ from NADH (Fig. 1 and S1†). HjLAD and SpNox were chosen for this study due to their high activity and stability. This system not only drives the thermodynamically unfavourable equilibrium of the reaction from L-arabinitol to L-xylulose, but also reduces the NADH inhibition present in the LAD reaction. In addition, the system offered high production yield at an initial substrate concentration of 250 mM by optimization of production parameters using response surface methodology (RSM).
 | ||
| Fig. 1 A schematic illustration of biocatalytic L-xylulose production by HjLAD coupled with SpNox, a cofactor regeneration enzyme. | ||
2. Materials and methods
2.1. Materials
Reagents for PCR and Ex-Taq DNA polymerase were purchased from Takara (Takara Shuzo Co., Japan). Restriction enzymes were obtained from New England Biolabs (Ipswich, MA, USA). pET-28a expression vector and plasmid isolation kit were from Qiagen (Hilden, Germany). Oligonucleotide primers were obtained from Bioneer (Daejeon, South Korea). L-Arabinitol, NADH, and NAD+ were obtained from Sigma-Aldrich (St. Louis, MO, USA). Electrophoresis reagents were provided by Bio-Rad (Hercules, CA, USA). Unless otherwise indicated, all chemicals were purchased from Sigma (St. Louis, MO, USA).2.2. Bacterial strains and culture conditions
The plasmids containing the wild-type SpNox gene25 and HjLAD gene26,27 were used for the production of SpNox and HjLAD proteins. The recombinant plasmids were transformed into Escherichia coli BL21 (DE3) for protein expression. E. coli strains harboring SpNox or HjLAD genes for protein expression were cultured at 37 °C in Luria–Bertani medium supplemented with kanamycin (50 μg mL−1). Isopropyl-β-D-thiogalactopyranoside (IPTG) was then added to the culture medium at a final concentration of 0.1 mM, and incubation continued with shaking at 25 °C. The induced cells were harvested by centrifugation at 4 °C for 15 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, rinsed with phosphate-buffered saline, and stored at −20 °C.
000 × g, rinsed with phosphate-buffered saline, and stored at −20 °C.
2.3. Preparation of cell-free extracts and protein purification and quantification
Cell pellets of SpNox were suspended in 50 mM potassium phosphate buffer (pH 7.0). Cell pellets of HjLAD were suspended in 100 mM Tris–HCl buffer (pH 8.0). The cell suspension was incubated on ice for 30 min in the presence of 1.0 mg mL−1 lysozyme. Cell disruption was carried out by sonication at 4 °C for 5 min, and the lysate was centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 20 min at 4 °C to remove cell debris, and the supernatant was used as the cell-free extract. Protein purification was processed as described elsewhere.25 The recombinant and purified proteins were analyzed by 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and visualized by staining with Coomassie Blue R-250. Protein concentration was determined by the Bradford method using bovine serum albumin as a standard protein.28
000 × g for 20 min at 4 °C to remove cell debris, and the supernatant was used as the cell-free extract. Protein purification was processed as described elsewhere.25 The recombinant and purified proteins were analyzed by 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and visualized by staining with Coomassie Blue R-250. Protein concentration was determined by the Bradford method using bovine serum albumin as a standard protein.28
2.4. Enzyme assay
Cell-free extracts of the recombinant strains were used for enzymatic activity assays. The activity of Nox and dehydrogenase were determined spectrophotometrically by monitoring the change in absorbance at 340 nm (A340) upon oxidation or reduction of NADH. The Nox assay mixture consisted of 200 μM NADH, 50 mM potassium phosphate buffer (pH 7.0), and enzyme in limiting amounts. The assay was carried out at room temperature. The HjLAD assay mixture for oxidation consisted of 0.5 mM NAD+, 200 mM L-arabinitol, and enzyme in 100 mM Tris–glycine–NaOH (pH 9.5). One unit corresponds to the oxidation of 1 μmol NADH or producing 1 μmol NADH per minute. All assays were performed in triplicate at least two separate times. The data represent the average of all statistically relevant data with a standard deviation of less than 10%.2.5. L-Sugar production assay
Activity of SpNox in cofactor regeneration was assayed with L-arabinitol dehydrogenase (LAD).26 All the reactions were run at room temperature with the standard buffer 100 mM Tris–HCl buffer, pH 8.0, in a total volume of 1 mL. The initial concentrations of L-arabinitol and NAD+ were 100 mM and 2 mM, respectively. Enzyme levels in the reaction solution were 0.1 mg SpNox variants and 0.36 mg LAD. A control reaction mixture containing only 0.36 mg LAD was processed similarly to those described above. One hundred-microliter samples were taken periodically to check the concentration of generated L-xylulose. The samples were analyzed by high-pressure liquid chromatography (HPLC) Ultimate 3000 (Dionex, CA, USA) equipped with a Shodex sugar SP0810 column (Showa Denko, K. K., Kawasaki, Japan) and an evaporation light scattering detector (ESA6700, MA). The elution buffer was water at a rate of 1 mL min−1 and the elution temperature was 80 °C (column temperature), while the evaporation and nebulizer temperatures were 50 °C and 65 °C, respectively. The retention times of L-arabinitol and L-xylulose were 9.8 and 16.4 min at the operation conditions, respectively. Quantification of L-xylulose derived from the reaction was done using external calibration standards with five different concentrations of L-xylulose. Each reaction was performed and analyzed at least twice and reported values are the average of the two measurements with the associated standard deviation.2.6. NADH inhibition
By using different concentrations of NADH as inhibitor, a series of kinetic measurements was performed at saturating constant concentrations of L-arabinitol with varying concentrations of the cofactor NAD+. The effect of NADH accumulation on L-arabinitol dehydrogenase kinetics was evaluated at initial NADH concentrations of 0, 50, 100 and 200 μM (duplicate experiments at each concentration). Identification of the type of inhibition was done by double-reciprocal analysis. The corresponding inhibition constants were derived from a non-linear fitting procedure using SigmaPlot 5.0 (Systat Software, San Jose, CA).2.7. Measurement of pyridine nucleotides in the reaction
Nucleotides were separated and analyzed by HPLC with a ZORBAX SB-C18 reverse-phase column (3.0 × 150 mm, 3.5 μm) (Agilent Technologies, Palo Alto, CA) with a UV detector (Waters model 486). NAD+ and NADH separated in isocratic conditions at 2.5% methanol in 0.1 M sodium phosphate buffer (pH 7.0). Standard curves were generated with authentic NAD+ and NADH so as to correlate peak area with cofactor amount. The flow rate was 1 mL min−1 at room temperature, and detection was at 254 nm.2.8. Response surface methodology
Factorial, central composite rotary design (CCRD) was used for four factors, with replicates at the center point and star points. The variables were HjLAD amount, NAD+ concentration, pH and temperature at each of 5 coded levels (−∞, −1, 0, +1, +∞). The actual levels of variables for the CCRD experiments were selected based on initial levels at the center points. A total of 30 experimental trials that included 16 trials for factorial design, 8 trials for axial points (2 for each variable), and 6 trials for replication of the central points were performed. The response value, which is the conversion (Y), was defined as the average of triplicates. Data were analyzed using Design-Expert software (Stat-Ease, Inc., Minneapolis, USA), including analysis of variance (ANOVA) to determine interactions between variables and responses.3. Results and discussion
3.1. L-Xylulose production by HjLAD in a cell-free system
Biocatalytic oxidoreductions are important in asymmetric synthesis, and have been receiving increasing attention in the enantiopure chemicals and pharmaceuticals industry. HjLAD, belonging to the polyol dehydrogenases (PDH), has been reported to catalyze the conversion of L-arabinitol to L-xylulose with concomitant NAD+ reduction in fungal L-arabinose catabolism. The turnover number (kcat) and the catalytic efficiency (kcat/Km) were 4200 min−1 and 290 mM−1 min−1, respectively.29,30 It is an important enzyme in the development of recombinant organisms that convert L-arabinitol to fuels and chemicals. We compared the biotransformation yield of L-arabinitol into L-xylulose using cell-free crude HjLAD and purified HjLAD. The conversion yield obtained with purified HjLAD and cell-free HjLAD using 20 mM L-arabinitol and 1 mM NAD+ were 3.1% and 53.1% in one hour, respectively. Compared to the conversion of the reaction using purified HjLAD, the same protein amount of cell-free HjLAD exhibited a significantly higher production of L-xylulose. In addition to HjLAD, it is likely that there are other enzymes present in the cell-free system involved in NAD+ regeneration that help facilitate the reaction towards L-xylulose production. In vitro biocatalytic cell-free systems have been studied well and utilized in many applications like the production of (3R)-acetoin from glycerol,31 production of dihydrogen from xylose,32 and transformation of nonfood biomass to starch.33 It shows advantages in higher product yields, faster reaction rates, and elimination of cell-associated process barriers compared to in vivo biocatalytic systems.34,35 It also exhibits economic benefits as there is no need to purify proteins. These observations motivated us to proceed with the production of L-xylulose using cell-free crude enzymes instead of purified enzymes.3.2. LAD inhibition by NADH
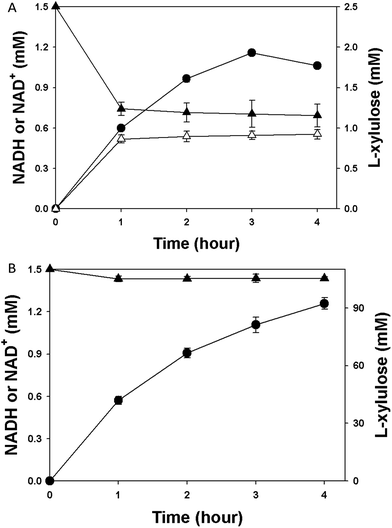
Several studies showed that PDHs suffer from NADH inhibition.19,24 In an attempt to gain insight into the inhibitory effects by the product NADH, the inhibition constant (Ki) of NADH for HjLAD was determined. Double reciprocal plots demonstrated a competitive inhibition of HjLAD by NADH (Fig. 2). The Ki value of NADH was determined to be 98.3 μM. SpNox catalyzes the oxidation of NADH by reducing molecular O2 to H2O with a Km of 27.0 μM and kcat/Km of 1.1 × 107 s−1 M−1. Not all the tested metals could enhance Nox activity, suggesting that the enzyme does not require a metal as cofactor for its activity.25 In order to address the inhibitory effect of NADH, the concentration profile of NADH in the reactions using purified HjLAD coupled or uncoupled with SpNox was determined. While 0.5 mM NADH was obtained in the reaction using cell-free HjLAD alone (Fig. 3A), NADH was not detected in the reactions using HjLAD coupled with SpNox (Fig. 3B). These results demonstrate that the decline in reaction velocity during biotransformation of L-arabinitol into L-xylulose is a result of the inhibition by the reaction product NADH. Therefore, it is vital to eliminate the NADH inhibition for efficient L-xylulose production.3.3. L-Xylulose production by coupling HjLAD and SpNox
HjLAD catalyzes strict NAD+-dependent interconversion between L-arabinitol and L-xylulose and exhibits NADH product inhibition. In order to make such a reaction more efficient and economical, it is necessary to regenerate the cofactor NAD+ from NADH. In the present work, we used the highly active SpNox for regeneration of NAD+. In the coupled system, SpNox was used to remove the reduced cofactor NADH from the reaction mixture and to recycle it to the oxidized cofactor NAD+. The cell-free HjLAD coupled with SpNox yielded 79.8% conversion of 20 mM L-arabinitol into L-xylulose in one hour. SpNox coupling with HjLAD in the reaction prevented HjLAD product inhibition by NADH, and shifted the reaction equilibrium towards the L-rare sugar product, and thereby could obtain a higher conversion compared to using cell-free HjLAD alone (53.1%).To explore the applications of the HjLAD–SpNox coupled enzyme system at higher concentrations of L-arabinitol, the effect of different concentrations of L-arabinitol on the conversion was further studied. When L-arabinitol conversion catalyzed by HjLAD was investigated at various concentrations of L-arabinitol and 3 mM NAD+ (Fig. 4), 90.7% and 71.6% yields were obtained after 8 hours with 150 mM and 200 mM L-arabinitol, respectively (Fig. 4). Biotechnological production of L-xylulose from previous studies mostly used the resting cell as biocatalyst.5,9,15–17 This is a higher yield of L-xylulose production compared to the previous reports at higher concentration of substrate.
In our previous study, the E. coli whole-cell biocatalysts could not achieve a high yield at higher (>150 mM) L-arabinitol concentrations.36 It might be due to the difficulty of substrate diffusion and the high communication barrier between two E. coli cells. However, a cell-free system, which leaves out the cell and removes cell-wall associated diffusion limitation, performed better in L-xylulose production at higher substrate concentration. In this study, 59.3% conversion was obtained when 250 mM L-arabinitol was used in the cell-free reaction system without any optimization step. Therefore, RSM was performed next to optimize the reaction conditions for L-xylulose production using 250 mM L-arabinitol.
3.4. RSM optimization of production conditions
To enhance the production of L-xylulose using 250 mM L-arabinitol, RSM optimization was performed. RSM has advantages over conventional statistical techniques by virtue of the fact that it investigates the effects of all variable parameters simultaneously. On the basis of initial L-xylulose production results, a statistical experimental design CCRD was employed to investigate the effect of four independent variables (HjLAD amount, NAD+ amount, pH, and temperature) on the production of L-xylulose using an excess amount of SpNox (6.25 U mL−1) (Table 1). Thirty experiments were performed to optimize these parameters as shown in Table 2. Among them, five replications were at center points (n = 5). The application of RSM yielded a regression equation, which was the empirical relationship between L-xylulose production and the test variables in coded units. The statistical significance of the model equation was evaluated by the F-test for analysis of variance (ANOVA), which showed that the regression was statistically significant. The ‘Prob > F’ value for the model was <0.0001, which indicated that the model was statistically significant with a confidence interval of 99.99%.| Variable | Symbol | Coded level | ||||
|---|---|---|---|---|---|---|
| −α | −1 | 0 | +1 | +α | ||
| HjLAD (U mL−1) | A | 1.37 | 2.74 | 4.12 | 5.49 | 6.86 |
| NAD+ (mM) | B | 2.5 | 5.0 | 7.5 | 10.0 | 12.5 |
| pH | C | 6 | 7 | 8 | 9 | 10 |
| Temperature (°C) | D | 20 | 25 | 30 | 35 | 40 |
| Run | HjLAD (U mL−1) | NAD+ (mM) | pH | Temperature (°C) | Conversion (%) | Predicted conversion (%) |
|---|---|---|---|---|---|---|
| 1 | 4.12 | 7.5 | 8 | 30 | 76.94 | 76.94 |
| 2 | 5.49 | 5 | 9 | 25 | 63.71 | 65.87 |
| 3 | 4.12 | 12.5 | 8 | 30 | 70.74 | 73.07 |
| 4 | 2.75 | 5 | 7 | 25 | 61.39 | 61.01 |
| 5 | 2.75 | 10 | 7 | 25 | 70.36 | 71.04 |
| 6 | 2.75 | 5 | 9 | 35 | 50.00 | 50.52 |
| 7 | 5.49 | 10 | 9 | 25 | 72.04 | 69.33 |
| 8 | 5.49 | 10 | 7 | 35 | 68.87 | 68.53 |
| 9 | 4.12 | 7.5 | 8 | 30 | 76.94 | 76.94 |
| 10 | 2.75 | 5 | 7 | 35 | 63.33 | 65.03 |
| 11 | 5.49 | 10 | 9 | 35 | 69.07 | 70.02 |
| 12 | 5.49 | 10 | 7 | 25 | 62.16 | 62.22 |
| 13 | 2.75 | 10 | 9 | 25 | 64.29 | 64.07 |
| 14 | 4.12 | 7.5 | 8 | 30 | 76.94 | 76.94 |
| 15 | 4.12 | 7.5 | 8 | 30 | 76.94 | 76.94 |
| 16 | 4.12 | 7.5 | 6 | 30 | 67.52 | 66.37 |
| 17 | 2.75 | 10 | 7 | 35 | 73.62 | 72.04 |
| 18 | 2.75 | 5 | 9 | 25 | 52.80 | 52.12 |
| 19 | 4.12 | 7.5 | 8 | 30 | 76.94 | 76.94 |
| 20 | 5.49 | 5 | 9 | 35 | 71.26 | 69.57 |
| 21 | 2.75 | 10 | 9 | 35 | 61.39 | 59.46 |
| 22 | 4.12 | 7.5 | 8 | 30 | 76.94 | 76.94 |
| 23 | 4.12 | 2.5 | 8 | 30 | 64.48 | 62.59 |
| 24 | 4.12 | 7.5 | 8 | 40 | 60.00 | 60.57 |
| 25 | 5.49 | 5 | 7 | 35 | 69.19 | 70 |
| 26 | 4.12 | 7.5 | 8 | 20 | 56.00 | 55.87 |
| 27 | 1.37 | 7.5 | 8 | 30 | 63.32 | 64.04 |
| 28 | 6.86 | 7.5 | 8 | 30 | 74.56 | 74.27 |
| 29 | 5.49 | 5 | 7 | 25 | 59.77 | 60.68 |
| 30 | 4.12 | 7.5 | 10 | 30 | 57.38 | 58.97 |
The ‘Model F-value’ of 43.2 also implied that the model was significant and that there was only a 0.01% chance that the ‘Model F-value’ could occur because of noise. Fig. 5 shows the isoresponse contour and surface plots for the optimized conditions of L-xylulose production. The effect of enzyme and NAD+ concentrations on L-xylulose production is shown in Fig. 5A. The production of L-xylulose increased with increasing NAD+ concentration. A high HjLAD concentration (4.9 U mL−1) with an NAD+ concentration of 8.5 mM resulted in improved conversion of L-xylulose. Fig. 5B shows the relationship between pH and enzyme concentration. At a fixed amount of SpNox present in the reaction, the conversion of L-xylulose increases up to around 76% as pH increases from 7.0 to 8.0 and HjLAD concentration increases from 2.74 U mL−1 to 4.6 U mL−1. Then the conversion of L-xylulose drops as both factors increase. The optimal pH for HjLAD and SpNox is 9.5 and 7.0, respectively.25,26 Reaction pH is a critical factor for both enzymes functioning well in the cell-free coupling system. Fig. 5C shows the effect of temperature and enzyme concentration on conversion. The lower temperature in the reaction might slow reaction, resulting in lower conversion, but higher temperature will disable enzyme function. Stability studies show that SpNox and HjLAD have a half-life (t1/2) of 5.8 h and 165 h at 30 °C, respectively.37,38 At higher temperatures, SpNox activity may be disabled and the overall reaction conversion reduced.
3.5. Validation and confirmation of the model
Generally, it is important to validate the fitted model to ensure that it sufficiently approximates the results obtained in the field conditions. An unfit model is very likely to give poor and misleading results. The coefficient of multiple regression, R2, is a global statistic for assessing the fit of a model. In the present model, the coefficient of determination R2 was 0.9758, which further indicated that the model was suitable for adequately representing the real relationships among the selected reaction variables. For further validation of the model, adjusted R2 was used for confirming model adequacy. The adjusted R2 was calculated to be 0.9532, which indicates a reasonably good model for the field conditions (Table 3). Numerical optimization using the CCRD was employed to predict the optimal levels of HjLAD concentration, NAD+ amount, pH, and temperature for maximal production of L-xylulose. Based on this method, a maximum conversion yield of 78.2% was predicted with an HjLAD level of 4.9 U mL−1, 8.2 mM of NAD+, pH 8.0, and temperature of 30.9 °C. The results using the optimized conditions were verified by carrying out experiments that incorporated the optimized variables. A maximum conversion efficiency of 78.4% was obtained, which was in close agreement with the model prediction. Fig. 6 shows the time course of the production of L-xylulose using 250 mM L-arabinitol.| Source | Mean square | F-Value | Prob > F |
|---|---|---|---|
| Model | 119.8443 | 43.19861 | <0.0001 |
| A – HjLAD | 156.962 | 56.57791 | <0.0001 |
| B – NAD+ | 164.6702 | 59.35638 | <0.0001 |
| C – pH | 82.19612 | 29.6281 | <0.0001 |
| D – temperature | 33.1914 | 11.96405 | 0.0035 |
| AB | 72.004 | 25.95428 | 0.0001 |
| AC | 198.1065 | 71.4087 | <0.0001 |
| AD | 28.14073 | 10.1435 | 0.0062 |
| BC | 3.681817 | 1.327134 | 0.2673 |
| BD | 9.03061 | 3.255139 | 0.0913 |
| CD | 31.53984 | 11.36873 | 0.0042 |
| A2 | 103.9643 | 37.47456 | <0.0001 |
| B2 | 142.3502 | 51.31101 | <0.0001 |
| C2 | 349.379 | 125.9358 | <0.0001 |
| D2 | 601.1807 | 216.6993 | <0.0001 |
Several previous studies on L-xylulose production have been conducted using resting cells as biocatalysts. However, low conversion efficiency was noted when high substrate concentration was used in the production reactions. Poonperm et al. reported an L-xylulose yield of 85% after 12 h of reaction with B. pallidus Y25 using 2% (∼131 mM) xylitol.9 A yield of approximately 53% was reported when resting E. coli cells harboring the recombinant xylitol dehydrogenase gene from B. pallidus were used to produce L-xylulose from 2% xylitol within 24–48 h.5 In the present study, a high conversion rate was obtained using 250 mM L-arabinitol within 8 h. This showed that cell-free systems can convert the substrate into product more efficiently than resting cells. Currently, we are working on the immobilization of crude SpNox and HjLAD for reuse as biocatalysts.
4. Conclusions
In conclusion, we successfully applied HjLAD coupled with SpNox for the regeneration of NAD+ to obtain a high L-xylulose yield from L-arabinitol. The present study clearly demonstrates that it is important to eliminate the reduced form of the pyridine nucleotide NADH, and to provide a sufficient amount of the oxidized form of pyridine nucleotide cofactors for NAD+-dependent dehydrogenation of L-arabinitol to L-xylulose. Coupling SpNox with polyol dehydrogenases including HjLAD in a biocatalytic cell-free system provides an efficient system for the production of various L-rare sugars including L-xylulose. In the future, protein engineering could be applied to improve the properties of SpNox and HjLAD to obtain more ideal biocatalysts for use in a cell-free enzyme system.Acknowledgements
This work was supported by the Department of Chemical Engineering and MCubed at the University of Michigan. This work was also supported by the Energy Efficiency & Resources Core Technology Program of the Korea Institute of Energy Technology Evaluation and Planning (KETEP), granted financial resource from the Ministry of Trade, Industry & Energy, Republic of Korea (201320200000420, 20153010092130, and 20153030091450). This work was supported by WTU Joint Research Grants of Konkuk University.Notes and references
- W. Poonperm, G. Takata and K. Izumori, Biosci., Biotechnol., Biochem., 2008, 72, 231–235 CrossRef CAS PubMed.
- K. Beerens, T. Desmet and W. Soetaert, J. Ind. Microbiol. Biotechnol., 2012, 39, 823–834 CrossRef CAS PubMed.
- Y. W. Zhang, M. K. Tiwari, M. Jeya and J. K. Lee, Appl. Microbiol. Biotechnol., 2011, 90, 499–507 CrossRef CAS PubMed.
- C. Mathé and G. Gosselin, Antiviral Res., 2006, 71, 276–281 CrossRef PubMed.
- G. Takata, W. Poonperm, K. Morimoto and K. Izumori, Biosci., Biotechnol., Biochem., 2010, 74, 1807–1813 CrossRef CAS PubMed.
- S. S. Jagtap, S. S. Dhiman, T.-S. Kim, I.-W. Kim and J.-K. Lee, Appl. Microbiol. Biotechnol., 2014, 98, 661–669 CrossRef CAS PubMed.
- R. Verho, M. Putkonen, J. Londesborough, M. Penttila and P. Richard, J. Biol. Chem., 2004, 279, 14746–14751 CrossRef CAS PubMed.
- Q. Meng, T. Zhang, B. Jiang, W. Mu and M. Miao, Appl. Microbiol. Biotechnol., 2016, 100, 535–540 CrossRef CAS PubMed.
- W. Poonperm, Enzyme Microb. Technol., 2007, 40, 1206–1212 CrossRef CAS.
- B. Seiboth, L. Hartl, M. Pail and C. P. Kubicek, Eukaryotic Cell, 2003, 2, 867–875 CrossRef CAS PubMed.
- H. Zhao and W. A. van der Donk, Curr. Opin. Biotechnol., 2003, 14, 583–589 CrossRef CAS PubMed.
- R. Singh, R. K. Singh, S.-Y. Kim, S. Sigdel, J.-H. Park, J.-H. Choi, I.-W. Kim and J.-K. Lee, Biochem. Eng. J., 2016, 109, 189–196 CrossRef CAS.
- R. Singh, R. K. Singh, I.-W. Kim, S. Sigdel, V. C. Kalia, Y. C. Kang and J.-K. Lee, Enzyme Microb. Technol., 2016, 72, 56–64 CrossRef PubMed.
- W. Liu and P. Wang, Biotechnol. Adv., 2007, 25, 369–384 CrossRef CAS PubMed.
- J. S. Aarnikunnas, A. Pihlajaniemi, A. Palva, M. Leisola and A. Nyyssola, Appl. Environ. Microbiol., 2006, 72, 368–377 CrossRef CAS PubMed.
- T.-S. Kim, H.-M. Jung, S.-Y. Kim, L. Zhang, J. Li, S. Sigdel, J.-H. Park, J.-R. Haw and J.-K. Lee, J. Microbiol. Biotechnol., 2015, 25, 1093–1100 CrossRef CAS PubMed.
- A. R. Khan, H. Tokunaga, K. Yoshida and K. E. N. Izumori, J. Ferment. Bioeng., 1991, 72, 488–490 CrossRef CAS.
- C. F. B. Witteveen, F. Weber, R. Busink and J. Visser, Microbiology, 1994, 140, 1679–1685 CrossRef CAS.
- R. Lunzer, Y. Mamnun, D. Haltrich, K. D. Kulbe and B. Nidetzky, Biochem. J., 1998, 336, 91–99 CrossRef CAS PubMed.
- B. Nidetzky, H. Helmer, M. Klimacek, R. Lunzer and G. Mayer, Chem.-Biol. Interact., 2003, 143–144, 533–542 CrossRef CAS PubMed.
- X. P. Yang, L. J. Wei, J. B. Ye, B. Yin and D. Z. Wei, Arch. Biochem. Biophys., 2008, 477, 206–210 CrossRef CAS PubMed.
- P. Ödman, W. B. Wellborn and A. S. Bommarius, Tetrahedron: Asymmetry, 2004, 15, 2933–2937 CrossRef.
- D. Schwartz, M. Stein, K.-H. Schneider and F. Giffhorn, J. Biotechnol., 1994, 33, 95–101 CrossRef CAS.
- M. Slatner, B. Nidetzky and K. D. Kulbe, Biochemistry, 1999, 38, 10489–10498 CrossRef CAS PubMed.
- H. Gao, M. K. Tiwari, Y. C. Kang and J.-K. Lee, Bioorg. Med. Chem. Lett., 2012, 22, 1931–1935 CrossRef CAS PubMed.
- M. K. Tiwari, R. K. Singh, H. Gao, T. Kim, S. Chang, H. S. Kim and J. K. Lee, Bioorg. Med. Chem. Lett., 2014, 24, 173–176 CrossRef CAS PubMed.
- M. Pail, T. Peterbauer, B. Seiboth, C. Hametner, I. Druzhinina and C. P. Kubicek, Eur. J. Biochem., 2004, 271, 1864–1872 CrossRef CAS PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- P. Richard, J. Londesborough, M. Putkonen, N. Kalkkinen and M. Penttila, J. Biol. Chem., 2001, 276, 40631–40637 CrossRef CAS PubMed.
- M. K. Tiwari, R. K. Singh, R. Singh, M. Jeya, H. Zhao and J. K. Lee, J. Biol. Chem., 2012, 287, 19429–19439 CrossRef CAS PubMed.
- C. Gao, Z. Li, L. Zhang, C. Wang, K. Li, C. Ma and P. Xu, Green Chem., 2015, 17, 804–807 RSC.
- J. S. Martín del Campo, J. Rollin, S. Myung, Y. Chun, S. Chandrayan, R. Patiño, M. W. W. Adams and Y. H. P. Zhang, Angew. Chem., Int. Ed., 2013, 52, 4587–4590 CrossRef PubMed.
- C. You, H. Chen, S. Myung, N. Sathitsuksanoh, H. Ma, X. Z. Zhang, J. Li and Y. H. Zhang, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 7182–7187 CrossRef CAS PubMed.
- S. Billerbeck, J. Harle and S. Panke, Curr. Opin. Biotechnol., 2013, 24, 1037–1043 CrossRef CAS PubMed.
- Y. H. Zhang, J. Sun and J. J. Zhong, Curr. Opin. Biotechnol., 2010, 21, 663–669 CrossRef CAS PubMed.
- H. Gao, I.-W. Kim, J.-H. Choi, E. Khera, F. Wen and J.-K. Lee, Biochem. Eng. J., 2015, 96, 23–28 CrossRef CAS.
- H. Gao, M. Tiwari, R. Singh, B. Sung, S. Kim and J.-K. Lee, Appl. Microbiol. Biotechnol., 2014, 98, 7081–7088 CrossRef CAS PubMed.
- R. K. Singh, M. K. Tiwari, R. Singh, J. R. Haw and J. K. Lee, Appl. Microbiol. Biotechnol., 2014, 98, 1095–1104 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra11614k |
| This journal is © The Royal Society of Chemistry 2016 |