Identification of new p300 histone acetyltransferase inhibitors from natural products by a customized virtual screening method†
Abstract
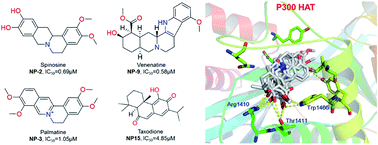
Herein, we sought to discover new p300 HAT inhibitors from natural products by a customized structure-based virtual screening method. The natural compounds NP-2 (spinosine), NP-3 (palmatine), NP-9 (venenatine), and NP-15 (taxodione) were found to be potent p300 HAT inhibitors, of which IC50 values are 0.69 μM, 1.05 μM, 0.58 μM, 4.85 μM, respectively.

 Please wait while we load your content...
Please wait while we load your content...