The effect of structural heterogeneity on the conformation and stability of Aβ–tau mixtures†
Abstract
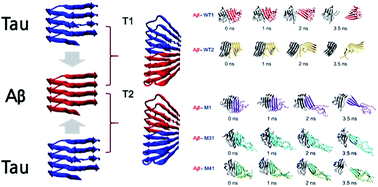
Oligomeric and fibrillar amyloids, which cause neurodegenerative diseases, are typically formed through repetitive fracture and elongation processes involving single homogeneous amyloid monomers. However, experimental and computational methods have shown that the amyloid proteins could be composed of heterogeneous amyloid segments. Specifically, owing to the polymorphism of amyloids under physiological conditions, it is crucial to understand the structural characteristics of heterogeneous amyloids in detail by considering their specific mutations and polymorphic nature. Therefore, in this study we used atomistic simulations to reveal the various structural characteristics of heterogeneous amyloids, which are amyloids composed of amyloid beta (Aβ) and mutated tau proteins. Furthermore, we showed that the different characteristics and conformations of Aβ–tau mixtures are the cause of the different types of tau proteins based on Aβ segments. Interestingly, we found that valine and lysine residues have a significant impact on the structural conformation and stability of the heterogeneous Aβ–tau mixtures. We also showed that two types of binding are key to understanding the different binding features and mechanical reactions to tensile load. This study sheds light on the assembly features of heterogeneous Aβ–tau mixtures as neurodegenerative disease factors.

 Please wait while we load your content...
Please wait while we load your content...