Investigation of various fluorescent protein–DNA binding peptides for effectively visualizing large DNA molecules†
Abstract
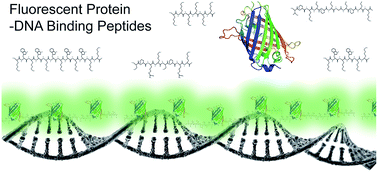
Large DNA molecules were visualized with novel fluorescent protein–DNA binding peptides (FP–DBPs). We constructed FP–DBPs by linking fluorescent protein to the N- or C-terminal of one or two peptides. We designed these DNA binding peptides from various DNA binding motifs such as oligo-lysine (K) and Lys-Trp (KW) repeats, TPKRPRGRPKK from high mobility group (HMG) chromosomal protein, and Ser-Pro-Arg-Lys (SPRK) from histone protein. We demonstrated the use of FP–DBP to stain large DNA molecules, and then analysed the fluorescence brightness and their binding affinity to double stranded DNA. This investigation provided HMG-tagged FP–DBP as the best DNA staining reagent in terms of fluorescence intensity, signal-to-noise ratio, and DNA binding affinity (Kd = 586 nM). Furthermore, we measured elongation of FP–DBP-stained DNA molecules tethered on the surface in order to evaluate FP–DBP-induced structural deformation.

 Please wait while we load your content...
Please wait while we load your content...