Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Detection of Listeria innocua on roll-to-roll produced SERS substrates with gold nanoparticles
S.
Uusitalo†
a,
M.
Kögler†
bg,
A.-L.
Välimaa
c,
A.
Popov
d,
Yu.
Ryabchikov
ei,
V.
Kontturi
f,
S.
Siitonen
f,
J.
Petäjä
a,
T.
Virtanen
h,
R.
Laitinen
c,
M.
Kinnunen
d,
I.
Meglinski
d,
A.
Kabashin
e,
A.
Bunker
b,
T.
Viitala
b and
J.
Hiltunen
*a
aVTT Technical Research Centre of Finland, Kaitoväylä 1, 90590 Oulu, Finland. E-mail: Jussi.hiltunen@vtt.fi
bCentre for Drug Research, Division of Pharmaceutical Biosciences, Faculty of Pharmacy of the University of Helsinki, Finland
cNational Resources Institute Finland (LUKE), Bio-based Business and Industry, University of Oulu, P. O. Box 413 (Paavo Havaksen Tie 3), FI-90014, Finland
dOptoelectronics and Measurement Techniques, Faculty of Information Technology and Electrical Engineering, University of Oulu, Finland
eLaboratoire Lasers, Plasmas Procédés Photoniques, Aix-Marseille University (AMU), 163 Avenue de Luminy, Case 917, 13288 Marseille Cedex 09, France
fNanocomp Oy Ltd, Ensolantie 6, 80710 Lehmo, Finland
gLaboratory of Bioprocess Engineering, Institute of Biotechnology, Technische Universität Berlin, Ackerstr. 71-76, D-13355 Berlin, Germany
hLappeenranta University of Technology, School of Engineering Science, Research Group of Membrane Technology, P. O. Box 20, FI-53851 Lappeenranta, Finland
iP. N. Lebedev Physical Institute of Russian Academy of Sciences, 53 Leninskii Prospekt, 119 991, Moscow, Russia
First published on 22nd June 2016
Abstract
The rapid and accurate detection of food pathogens plays a critical role in the early prevention of foodborne epidemics. Current bacteria identification practices, including colony counting, polymerase chain reaction (PCR) and immunological methods, are time consuming and labour intensive; they are not ideal for achieving the required immediate diagnosis. Different SERS substrates have been studied for the detection of foodborne microbes. The majority of the approaches are either based on costly patterning techniques on silicon or glass wafers or on methods which have not been tested in large scale fabrication. We demonstrate the feasibility of analyte specific sensing using mass-produced, polymer-based low-cost SERS substrate in analysing the chosen model microbe with biological recognition. The use of this novel roll-to-roll fabricated SERS substrate was combined with optimised gold nanoparticles to increase the detection sensitivity. Distinctive SERS spectral bands were recorded for Listeria innocua ATCC 33090 using an in-house build (785 nm) near infra red (NIR) Raman system. Results were compared to both those found in the literature and the results obtained from a commercial time-gated Raman system with a 532 nm wavelength laser excitation. The effect of the SERS enhancer metal and the excitation wavelength on the detected spectra was found to be negligible. The hypothesis that disagreements within the literature regarding bacterial spectra results from conditions present during the detection process has not been supported. The sensitivity of our SERS detection was improved through optimization of the concentration of the sample inside the hydrophobic polydimethylsiloxane (PDMS) wells. Immunomagnetic separation (IMS) beads were used to assist the accumulation of bacteria into the path of the beam of the excitation laser. With this combination we have detected Listeria with gold enhanced SERS in a label free manner from such low sample concentrations as 104 CFU ml−1.
Introduction
Foodborne diseases represent a serious public health issue. The incidence of epidemics related to food pathogens has increased significantly due to the greatly accelerated range and speed of distribution that has resulted from the increasingly global trade network for food products.1 For this reason, food safety authorities around the world have realized the need for a strict regulatory framework, including an exhaustive food testing regime.2,3 Traditional methods for the detection of bacteria, include direct culture and colony counting, polymerase chain reaction (PCR) and immunological methods. These are all extremely labour intensive and time consuming;1,4–6 the extent to which a rapid and efficient food testing regime can be achieved using the currently available methodologies is limited.Raman spectroscopy is a promising new methodology for bacteria detection, with many advantages including identification of the specific species of the bacteria, rapid detection, multiple simultaneous analyses and being label free.7–11 The identification of the species of bacteria through Raman spectroscopy is achieved through the detection of organic molecules on the surface of the bacterial membrane and wall. Their chemical structure provides a specific fingerprint of the bacterium that shows up in the Raman spectrum.7 The detection of a small concentration of bacteria with conventional Raman spectroscopy can demand a level of sensitivity greater than what it is capable of. The usual lowest detected bacterial concentration being around 108 CFU ml−1.9,10,12 Regulatory agencies demand finding a single cell in 25 g of food and such low concentrations require a brief pre-enrichment step to reach a more detectable level of 104 to 105 CFU ml−1.3 The conventional Raman spectroscopy cannot reach this level and the signal thus needs to be amplified. This can be achieved through the use of noble metallic materials, for example gold or silver, to trigger localised surface plasmons.13 Surface Enhanced Raman Scattering (SERS) is a special type of Raman spectroscopy, where irregular or patterned metal substrates or metal nanocolloids of different shape and size can be used for the signal enhancement.13–16
Typically the best enhancement effect is achieved with silver induced SERS.17 However, the use of silver has some drawbacks. As a substance it is antimicrobial and thus affects the sample under inspection. It is chemically quite reactive and the stability and reproducibility of the silver substrates and colloids (AgNP) can also be an issue.13,15 Gold is preferred in microbe detection as it is stable, non-toxic and has the optimal excitation wavelength in the near-infra red region, reducing auto-fluorescence issues generated by the microbes.
Among the foodborne pathogens Listeria monocytogenes is the most common culprit in causing death due to food poisoning. The fatality rate for L. monocytogenes infection is relatively high, ranging from 20 to 30%.4L. monocytogenes is an especially difficult pathogen to control, as a result of its tolerance to a wide range of temperatures and pH conditions. The detection of Listeria spp. with SERS has been studied previously with different SERS enhancers including different SERS substrate4,18 approaches and SERS colloid17,19–23 research. The SERS substrates have several advantages over the colloids; these include more consistent patterns without unforeseen aggregation and the ability to act as a base for the entire analysis chip. On the other hand, colloids can reach better sensitivity and be preferable when there is a need to detect features in the region of a larger organism like bacteria.
The direct detection of bacteria in food is difficult due to background signals and requires the disintegration of the solid food containing the bacteria, e.g. by mechanical methods, followed by culturing of the bacteria at elevated temperatures in the food matrix to accelerate the bacteria growth to a detectable level. Weidemaier et al.,24 has studied the detection of L. monocytogenes inside the food matrix with the help of immunomagnetic beads and nanoparticle SERS tags with antibodies, as they point out, the method is sensitive to the extent to which the magnetic particles can be concentrated within the area of the laser beam and care must be taken to succeed with reproducible pelleting of the magnetic particles. A more common method for bacteria detection is to remove the bacteria from the food matrix, often with immunocapture and subsequently pre-enrich the concentration before detection.6 Although the pre-enrichment technique requires culturing this can be used as a normalising factor for the state of the pathogen. The growth of Listeria at different temperatures, for example 4, 25 or 37 °C, produces bacteria with different amount of flagella and a different level of virulence. This affects the spectral fingerprint of the bacteria; in order to obtain reproducible spectra for bacteria, the same growing conditions in addition to detection processes are required.18,25
In previous research, it has been shown that it is possible to differentiate bacteria through SERS analysis.25–28 Preliminary results also indicate that SERS can be used for identifying bacteria and spores even at a strain level.18,25 Usually this requires a high concentration of bacteria. With low concentration the assistance of immunocapture may be needed for the separation of different bacteria. The genus Listeria consists of fifteen species from which only Listeria monocytogenes is pathogenic to humans.29 Immunocapture by current commercially available antibodies can seldom distinguish between the Listeria species. There are studies focussing on the production of high quality antibodies only for L. monocytogenes.30 The presence of non-pathogenic Listeria such as Listeria innocua may, however, indicate also contamination with L. monocytogenes.30 Furthermore, as the morphologic structure of L. innocua is similar to L. monocytogenes and their Raman/SERS-spectra are quite similar, L. innocua can be used as a model for Listeria detection.20
The detection of Listeria with SERS has been previously studied by several research groups. The majority of studies have focussed on the detection of Listeria at high concentration, 108 to 1010 CFU ml−1,4,17–19,22,23,30,31 and many have used chemometric analysis for the separation of Listeria spectra from the spectra of other pathogenic bacteria.4,17,19,31 Fewer studies have focussed on lowering the detection limit of the SERS procedure for Listeria detection than on the acquisition of representative spectra. Chen et al. have developed a method for detection of L. monocytogenes and L. innocua by in situ synthesis of silver nanoparticles.21 The limit of detection for the model sample L. innocua was found to be 103 CFU ml−1. The assay for bacteria detection was, however, performed for bacteria in pure water and required an extra incubation step with the silver colloids and two washing steps after the incubation.
The objective of this study was to develop a simplified and affordable method for label-free detection of Listeria with high sensitivity that is possible to perform on a structured SERS substrate. The conventional way for fabricating structured SERS substrates is to use methods such as spin-coating, dip coating, chemical vapour deposition, electrochemical synthesis, electron beam lithography and etching.32–35 However, they are not optimised for manufacturing single use chips in terms of through-put volume or cost. There are also many methods such as liquid–liquid interface formation, pulsed laser deposition on microscope slides and reduction of gold chloride III in natural rubber membranes which have been only tested in lab scale as batch fabrication.36–39 The fabricated sensor areas are often small and the fabrication methods are difficult to transfer into high volume production required of truly disposable sensor chips. Our approach is to fabricate the structured SERS substrates on polymer webs in large scale with UV imprint lithography. This enables the fabrication of large sensor surface areas which can be easily cut into smaller SERS substrates.40,41 The fabricated SERS substrates are coated with a thin layer of gold by evaporation before integration of hydrophobic sample wells. This SERS platform is suitable for low cost high volume production and is practical for one-time use, which diminishes the contamination issues often encountered in microbe detection. Gold colloids were added to gain additional plasmonic enhancement. The method uses immunomagnetic separation (IMS) beads as bacteria cell concentrators and the only washing steps occur during the pre-enrichment phase. SERS enhancement of different types of gold nanoparticles with Listeria was studied and the colloids with the best enhancement effect were used in combination of R2R nanostructured gold SERS substrates.
Experimental
Gold nanoparticle synthesis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 in H2O.
5 in H2O.
AgNPs with the average size of 40 nm were purchased from Sigma-Aldrich for reference measurements with the commercial time-gated Raman spectrometer.45
SERS substrate fabrication
The SERS patterns were imprinted on top of a poly(methyl methacrylate) (PMMA) polymer sheet with roll-to-roll UV-nanoimprint lithography.41 The produced polymer webs and die-cut sheets before and after gold deposition are presented in Fig. 1. Reverse gravure technique was used to apply UV-curable lacquer on top of the PMMA web. Embossing reel was used to imprint the SERS patterns and the lacquer was cured through the PMMA web with UV light exposure. After die-cutting SERS substrates from the roll, a 240 nm gold layer was added by evaporating on top of the polymer SERS surface.41 | ||
| Fig. 1 Photograph of roll-to-roll patterned polymer webs and die-cut sheets before and after gold deposition. | ||
PDMS well integration
Sample wells were created into 1 mm thick PDMS sheets (Wacker, Elastosil) by biopsy punches of a diameter of 1–2 mm. These PDMS wells were bonded onto the polymer SERS substrates by physical adsorption. The hydrophobicity of the wells forces the sample to retreat inside the PDMS well and have contact with the gold layered patterned SERS surface.Cultivation of L. innocua and IMS bead separation
L. innocua ATCC 33090 was cultivated in LEE Broth (Labema, Lab M Limited, pH 7.2 ± 0.2) at 35 °C for 20 h without shaking. The concentration was analysed spectrophotometrically (Dynamica HALO DB-20S) and diluted into concentration series (103 CFU ml−1 to 109 CFU ml−1) in LEE broth. IMS was performed using Dynabeads® anti-Listeria (Life Technologies (Invitrogen) 71006), and a Dynal Magnetic Particle Concentrator DynaMag™-2 (Invitrogen Dynal) as follows: 1 ml volumes of bacterial culture was added to each of the microcentrifuge tubes containing a 20 μl volume of Dynabeads® anti-Listeria (Dynal) followed by incubation at room temperature for 10 min with continuous mixing by Mix-Mate (Eppendorf). The beads were concentrated by magnetic field (in the Dynal MPC-M) onto the side of the tube for 3 min, supernatants were carefully aspirated and the samples were washed with the washing buffer (0.15 M NaCl, 0.01 M sodium phosphate buffer, pH 7.4 with 0.05% Tween 20). After that the beads were concentrated and the supernatant removed. Finally, the bead–bacteria complexes were resuspended into 100 ml of washing buffer for the SERS detection. For reference a concentration analysis was performed with 50 μl volumes of bead–bacteria complexes streaked onto differential selective agar Listeria acc. to Ottaviani and Agosti (ALOA) chromogenic agar (Labema) and incubated at 35 ± 0.5 °C for 24–48 h.SERS spectral acquisition of Listeria innocua and post-processing of data
Surface-enhanced Raman spectroscopy (SERS) spectra of L. innocua with AuNPs was detected from samples pipetted into PDMS wells integrated on top of SERS-active substrates. Sample amounts varied from 5 to 10 μl and well diameter varied from 1 to 2 mm. Bacteria samples were pipetted into the wells and the chosen AuNPs were pipetted sequentially. SERS spectra were recorded with an in-house built Raman system coupled into an Olympus microscope with a 785 nm continuous wave (cw) laser. The minimum laser power irradiation used was 10 mW with 40× magnification to excite the samples. A maximum of 40 mW was used in combination with low magnification (20×). The signal collection time was 5 seconds without averaging.Reference spectra for L. innocua were recorded with 40 nm sized AgNPs by a commercial 532 nm picosecond pulsed laser time-gated Raman spectrometer (TimeGate Instruments Oy, Finland) with an average power of 10 mW, as well coupled into Olympus microscope.45 The signal integration time was set to cover the SERS-signal and the fluorescence decay-time from 0.9–1.6 ns. The bacteria sample was pipetted on top of a glass slide prior to the detection. TimeGate measurements where analysed with TimeGate spectral processing tool and the acquired data was baseline corrected with a simple linear algorithm in Matlab (release 2015a, Mathworks Inc., USA) after opening the data with the PLS toolbox, version 2.0 (Eigenvector Research Inc., Manson, WA, USA). Further data handling and figure plotting was executed with Origin Pro (version 9.4, OriginLab corp., USA).
Results and discussion
Methods for the detection of Listeria innocua
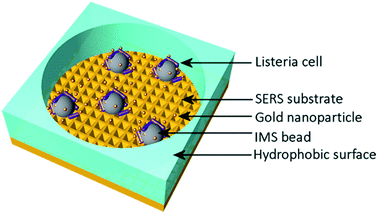
Typically Listeria spp. has been identified by the SERS method from highly concentrated samples, mostly in the range of 107 CFU ml−1 to 1010 CFU ml−1. When the detected concentrations are more realistic and the bacterial cells available for the detection are fewer, then the intensity of the detected Raman peaks diminishes and many of the peaks disappear from the spectrum. Thus it is more difficult to identify the bacteria from other bacterial species and the background with incomplete spectra. In these cases the identification of the bacteria can be handled by pathogen-capture proteins while SERS is used for the detection.29 Grow et al. have detected Listeria on planar SERS substrates by capturing and accumulating bacterial cells near the surface with an antibody layer.18 Although they concluded that the use of antibodies was possible and identification of bacteria was successful, the use of antibodies on the surface weakened the signal. This was assumed to be due to the increased separation distance between the surface and the bacterial cells.46 Thus, this approach with planar SERS substrates was not optimal. Another possibility for capturing bacteria is the use of immunomagnetic separation beads which have been used in several Escherichia coli studies34,35 and at least in one Listeria growth study.24 Usually, the IMS beads are either removed before detection or used as a part of a customised sandwich assay with SERS labels. In order to simplify the detection process we have developed a method to detect L. innocua in a label-free manner with IMS beads present during the SERS detection. The bacterial cells were captured by using a commercially available IMS separation kit. The sample was placed into the hydrophobic PDMS well on top of the polymer SERS substrate as is shown in Fig. 2 with the gold nanoparticles. Without IMS beads and PDMS well the bacterial cells in the liquid droplets spread wide apart and typically due to evaporation accumulated randomly to the droplet edges. We found that by using immunomagnetic separation beads during the detection we obtained a more stable SERS signal due to the more constant settling of the heavy IMS beads on to the sensor surface. A close-up transmission electron microscopy (TEM)picture of the IMS beads and a scanning electron microscopy (SEM) picture of IMS beads on top of the patterned SERS substrate can be seen in Fig. 3a and b respectively. In order to further enhance the SERS signal and to detect the features of the bacterial cells, gold nanoparticles were added around the bacteria bound to IMS beads.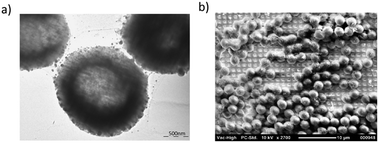 | ||
| Fig. 3 (a) TEM image of IMS beads with gold nanoparticles. The scale in the picture is 500 nm. (b) SEM image of the IMS beads (Invitrogen dynabeads) on top of patterned SERS substrate. | ||
AuNP characterisation
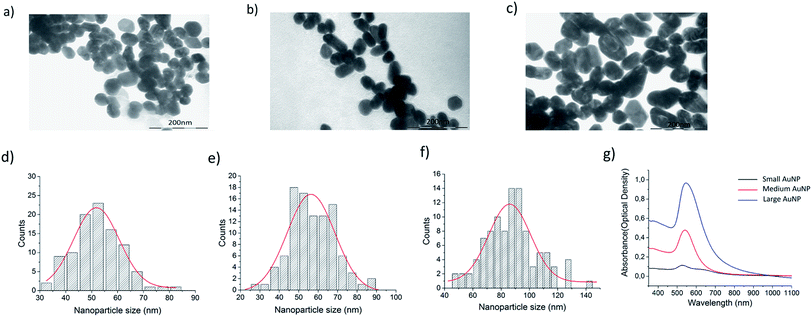
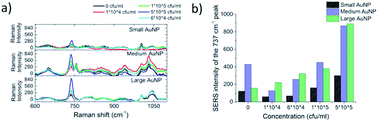
In search for the optimal gold nanoparticles (AuNPs) for bacteria detection with SERS, 3 candidates were selected. Ultrapure small AuNPs fabricated by femtosecond laser fragmentation were chosen as they could show better biocompatibility with bacteria cells than the synthesized AuNPs.42,47 The physically fabricated AuNPs lack the traces of non-reacted starting reagents, by-products, ions and surfactants, and have an additional advantage of lower background signal. Chemically synthesized medium size AuNPs and larger AuNPs were chosen to compare the signal intensity using differently shaped and sized particles. The NP size has been previously shown to matter in micro-organism detection and a rod like shape has seemed beneficial for SERS enhancement.48 The size and morphology of the fabricated AuNPs were retrieved by transmission electron microscopy (TEM) by using a LEO 912 OMEGA (Zeiss, Germany). One droplet of the 10 μl of aqueous nanostructure suspension was deposited onto a carbon-coated copper grid for TEM characterization. Fig. 4 shows the TEM images, the corresponding size distributions of the AuNP with Gaussian fit and the UV-Vis spectra of the AuNP. The maximum size for the different AuNP was estimated with the help of Gaussian fit shown in Fig. 4d–f. For the ultra-pure AuNPs the maximum size was found to be around 50 nm. The medium sized AuNPs showed a maximum of 60 nm with occasional large 90 nm sized particles. The large AuNPs had a maximum of 85 nm and a more rod like shape. From the UV-Vis spectra in Fig. 4g it can be seen how the maximum absorption peak of AuNPs shifts closer to 600 nm wavelength as the maximum size of the particles grows from 50 nm to 85 nm.The SERS effectiveness of the different sized AuNPs was studied by pipetting 5 μl of bacteria sample and 2 μl of concentrated NP solution into 2 PDMS wells positioned on top of the patterned SERS substrate. The acquired SERS spectra are presented in Fig. 5a. According to the bar plot of the intensity of 737 cm−1 peak presented in Fig. 5b, the medium size and the large size particles gave similar intensities for a bacteria concentration of 5 × 105 CFU ml−1. However, the large nanoparticles were chosen for further studies because the maximum of their UV-Vis spectra was closest to 785 nm and they provided more consistent spectra compared to the other AuNPs, which originated from the lower amount of background peaks thus giving a better resolution. These results strengthen the hypothesis that larger nanoparticles enhances the signal more than small round ones for microbe detection.49 The way the AuNPs were fabricated played a minor role in enhancing the signal. Physical ablation could not benefit the detection in such a manner which would have counterweighted the advantage of the size and the shape of the particles.
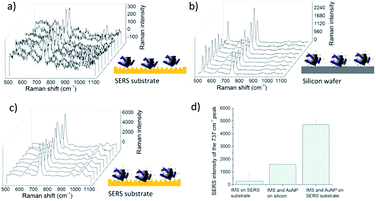
The development of the detection process
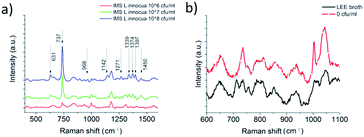
Commercial Dynabeads were used to capture the L. innocua cells for the SERS detection. The SERS signal recorded with IMS beads, possibly due to accumulation of more bacterial cells inside the excitation laser spot, was found to be 20 times stronger than the signal recorded without the beads. Fig. 6a shows the results for the comparison of studies with and without IMS beads and Fig. 6b shows the variation between 9 measurements points measured with IMS beads. The detection of L. innocua with the IMS beads was further studied on top of the patterned SERS substrate without AuNPs, as well as on top of silicon wafer with AuNPs and on top of patterned SERS substrate with AuNPs to see if there was an advantage in combining the SERS substrate with the AuNPs for bacteria detection. Fig. 7 represents the intensity differences between the measurements and it can be seen that the best intensities for the main dominant peak of 737 cm−1 were reached with the combination of the SERS substrate and AuNPs.The detected SERS lines for L. innocua ATCC 33090 and the effect of the traces of culture media and buffers on the SERS spectra
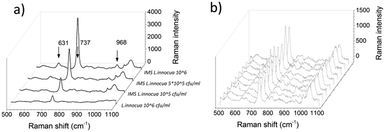
The captured L. innocua was first detected inside a PDMS well in a liquid state with a 40 mW laser power and a 20× magnification. Fig. 8a shows the mean spectra for the L. innocua specific Raman bands. It can be noted that when the bacterial amount diminishes some lines stay constant showing the lines created by the traces of culture medium and buffer liquids. Thus, it can be concluded that 9 Raman bands initiating from the bacterial cells were detected. Fig. 8b shows the Raman bands created by the sample matrix and by the original cultivation media of the bacterial cells, i.e. the LEE broth. Most of the background bands seem to originate from traces of the LEE broth.The nine Raman lines detected for L. innocua are listed in Table 1 with tentative assignments found in literature references. The dominant peak at 737 cm−1 has been previously suggested to originate from a glycosidic ring, adenine or CH2 rocking.50 Since the presence of adenine on the surface of the bacterial cell is unlikely and since the outer wall structure of Gram-positive bacteria such as Listeria spp. consists of a thick peptidoglycan structure rich in N-acetyl D-glucosamine (NAG), the origin of the peak is more likely caused by a glycosidic ring mode of NAG than adenine.23,51
| Detected lines | Raman shift (cm−1) | Tentative assignments | Reference |
|---|---|---|---|
| 631 | 627/620 | Phenylalanine (skeletal) | Luo, Lin 2008,23 Maquelin et al. 2002 (ref. 56) |
| 737 | 732 | Glycosidic ring mode of D-glucosamine (NAG), adenine or CH2 rocking | Luo, Lin 2008,52 Cui et al. 2015 (ref. 57) |
| 968 | 955 | N–C stretching | Vohník et al. 1998 (ref. 53) |
| 1142 | 1134/1130 | C–N and C–C stretch (carbohydrates) | Fan, Hu 2011,54 Chen et al. 2015 (ref. 58) |
| 1271 | 1230–1295 | Amide III | Liu, Chen 2007,59 Lu, Al-Qadiri 2011,60 Maquelin et al. 2002 (ref. 56) |
| 1339 | 1334/1339/1338 | Deformation CH/amide III/signature of adenosine monophosphate and guanosine monophosphate, aromatic amino acids tyrosine and tryptophan | Maquelin et al. 2000,61 Vohník et al. 1998,53 Harz, Rösch, Popp 2008 (ref. 62) |
| 1374 | 1371 | DNA | Harz, Rösch, Popp 2008 (ref. 62) |
| 1397 | 1392/1398 | Symmetric deformation of CH3 groups | Fan, Hu 2011,54 Al-Qadiri, Lin 2008 (ref. 55) |
| 1450 | 1453 | CH2 deformation (lipids) | Fan, Hu 2011,54 Cui et al. 2015 (ref. 57) |
The three closely aligned lines in the range of 1300–1400 cm−1 are interesting since three of them together have not been detected with L. innocua or L. monocytogenes in previous studies. The line 1339 cm−1 has been previously detected with Listeria by Luo et al.52 as a shifted line 1331 cm−1 which was suggested to originate from CH2 deformation. However, there are closer assignments to the detected 1339 cm−1 listed in E. coli studies. Vohník et al.53 have suggested that the exact line 1339 cm−1 could be originating from amide III and Harz et al. suggest that the line is due to the signature of adenosine monophosphate and guanosine monophosphate, aromatic amino acids tyrosine and tryptophan. Harz et al.7 also have a listing very near to the second line 1374 cm−1 assigned to DNA. The last line of the group 1397 cm−1 is most likely due to the symmetric deformation of CH3 group which has also been detected for the case of E. coli.54,55
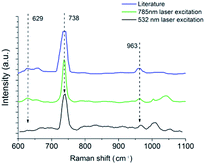
When comparing the Raman bands detected for L. innocua with the previous research, it is interesting to note that the SERS spectrum in different studies varies. Liu et al. among others has stated that this could be due to the differences in the measurement conditions such as the culture broth and temperature that have been used, excitation wavelength of the laser or the SERS enhancer.19,51 To test this hypothesis we recorded the SERS spectra of the same L. innocua sample with changed SERS conditions. Fig. 9 shows the peaks detected around the dominant peak 737 cm−1 for 3 different SERS conditions. In the first case the combination of AuNPs on top of the patterned SERS substrate with 785 nm cw excitation was used, while in the second case a SERS spectrum was detected from the same sample on top of a glass slide with AgNPs and pulsed laser excitation at a different wavelength of 532 nm. The results are consistent. They are also similar to the third case published by Luo et al. who used AgNPs with 785 nm cw excitation wavelength. Another research group of Kairyte et al.22 used silver NPs with 1064 nm excitation with a similar outcome. Clearly when comparing the results, there is no connection between the variations in spectra and the enhancer used (silver/gold). Additionally the excitation wavelength does not seem to affect the detected spectrum.
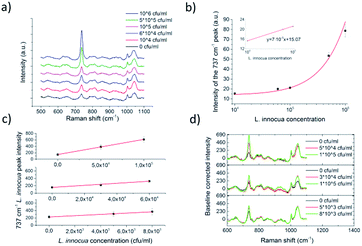
The detection process was developed further by manually lowering the minimum laser power of the Raman system to 10 mW. This enabled the use of larger magnification with the microscope without burning of the dried specimen during the measurement procedure. The sample density on the SERS substrate was also reduced to prevent the blocking of signal by the media traces of the sample liquids. It was noted that the media was disturbing the SERS signal if an excessive amount of traces had dried on top of the SERS substrate. Limit of detection and repeatability of the detection process could have been enhanced further with extra washing steps for the IMS beads, but this was avoided in order to not complicate the sample handling. Fig. 10 shows the mean intensity changes of the dominant peak of 737 cm−1 as a function of L. innocua concentration with 40× magnification and 10 mW laser excitation power for several concentration series. For the concentration series in Fig. 10a and b the Raman intensity was normalised by the background peak at 787 cm−1. The intensity of the dominant 737 cm−1 peak is displayed in Fig. 10b as a function of logarithmic L. innocua concentration that follows an exponential curve. For the concentration range below 105 CFU ml−1 the relation was found linear. Since the blank 0 CFU ml−1 sample exhibits a signal at 737 cm−1, the lowest limit of detection was considered through the deviation of the background signal generated by the sample matrix. According to the international union of pure and applied chemistry, IUPAC, the limit of detection (LOD) can be defined as the smallest concentration detected with reasonable certainty, and derived from
| LOD = ksbiS | (1) |
| S = Δc/ΔI,63 | (2) |
As a summary, we demonstrated in this study the use of disposable polymer SERS platforms and AuNPs with integrated sample wells for fast and simple detection of L. innocua. We have shown how the capture and deposit of the IMS bound bacteria cells onto the SERS substrate benefits the detection. In the future, the detection process could be further developed by utilizing the magnetic nature of the IMS beads on the SERS substrate for the removal of matrix traces e.g. by removing the matrix with wicking.
Conclusions
This study analyses the use of different types of AuNPs in addition to a structured polymer SERS substrate for Listeria detection. The polymer based SERS substrate has been produced with roll-to-roll fabrication and thus it is suitable for one time use due to the high volume production and the low cost per substrate. The results of this study provide new insights into Listeria diagnostics. We also demonstrate the benefit of using immunomagnetic separation beads as an accumulation assistant of the bacteria for enhanced signal intensity. The use of novel hydrophobic PDMS wells for sample preparation on SERS chips enables controlled sample appliance and reduces mean absolute deviation of SERS signals. The limit of detection in this methodology was determined to be in the range of ∼104 CFU ml−1 shown for the first time with label-free gold enhanced SERS using optimized AuNPs combined with an Au based SERS substrate.Acknowledgements
This project was funded by TEKES (the Finnish Funding Agency for Technology and Innovation) through FMA project and University of Oulu Graduate School through Infotech Oulu Doctoral Program and by Academy of Finland through FOULSENS (Grant No. 292253), M-SPEC (Grant No. 284907) and multi Diagnostics (Grant No. 290596). The financial support of the aforementioned institutes is gratefully acknowledged. We also thank Tiina Väyrynen (National Resources Institute Finland (LUKE)) for helping with the bacterial sample preparation. Yury Ryabchikov acknowledges a support from COST project (ECOST-STSM-BM1205-120416-072252) for performing experiments.Notes and references
- J. Law, N. A. Mutalib, K. Chan and L. Lee, Front Microbiol, 2013, 5, 770 Search PubMed.
- M. Gandhi and M. L. Chikindas, Int. J. Food Microbiol., 2007, 113, 1–15 CrossRef PubMed.
- U. Gasanov, FEMS Microbiol. Rev., 2005, 29, 851–875 CrossRef CAS PubMed.
- N. A. Mungroo, G. Oliveira and S. Neethirajan, Microchim. Acta, 2015, 183, 697–707 CrossRef.
- V. Velusamy, K. Arshak, O. Korostynska, K. Oliwa and C. Adley, Biotechnol. Adv., 2010, 28, 232–254 CrossRef CAS PubMed.
- X. Zhao, C. Lin, J. Wang and D. Oh, J. Microbiol. Biotechnol., 2014, 24, 297–312 CrossRef CAS PubMed.
- M. Harz, P. Rösch and J. Popp, Cytometry, Part A, 2009, 75, 104–113 CrossRef CAS PubMed.
- X. Lu, H. M. Al-Qadiri, M. Lin and B. A. Rasco, Food Bioprocess Technol., 2011, 4, 919–935 CrossRef.
- X. Lu, B. A. Rasco, D. H. Kang, J. M. F. Jabal, D. E. Aston and M. E. Konkel, Anal. Chem., 2011, 83, 4137–4146 CrossRef CAS PubMed.
- J. Wang, X. Xie, J. Feng, J. C. Chen, X. Du, J. Luo, X. Lu and S. Wang, Int. J. Food Microbiol., 2015, 204, 66–74 CrossRef CAS PubMed.
- I. Boyaci, H. Temiz and H. Geniş, RSC Adv., 2015, 5, 56606–56624 RSC.
- E. Smith and G. Dent, Modern Raman spectroscopy: a practical approach, 2013 Search PubMed.
- J. Anker, W. Hall, O. Lyandres and N. Shah, Nat. Mater., 2008, 7, 442–453 CrossRef CAS PubMed.
- S. Nie, Science, 1997, 275, 1102–1106 CrossRef CAS PubMed.
- K. Bantz, A. Meyer and N. Wittenberg, Phys. Chem. Chem. Phys., 2011, 13, 11551–11567 RSC.
- O. Bibikova, A. Popov, A. Bykov, A. Prilepskii, M. Kinnunen, K. Kordas, V. Bogatyrev, N. Khlebtsov, S. Vainio and V. Tuchin, J. Biomed. Opt., 2015, 20, 076017 CrossRef PubMed.
- C. Fan, Z. Hu, A. Mustapha and M. Lin, Appl. Microbiol. Biotechnol., 2011, 92, 1053–1061 CrossRef CAS PubMed.
- A. E. Grow, L. L. Wood, J. L. Claycomb and P. A. Thompson, J. Microbiol. Methods, 2003, 53, 221–233 CrossRef CAS PubMed.
- Y. Liu, Y.-R. Chen, X. Nou and K. Chao, Appl. Spectrosc., 2007, 61, 824–831 CrossRef CAS PubMed.
- G. Green and A. Chan, Instrumentation and Measurement Technology Conference Proceedings (IMTC '08), 2008, pp. 513–517 Search PubMed.
- L. Chen, N. Mungroo, L. Daikuara and S. Neethirajan, J. Nanobiotechnol., 2015, 13, 45 CrossRef PubMed.
- K. Kairyte, Z. Luksiene and V. Sablinskas, Chem. Technol., 2012, 61, 46–49 Search PubMed.
- B. S. Luo and M. Lin, J. Rapid Methods Autom. Microbiol., 2008, 16, 238–255 CrossRef CAS.
- K. Weidemaier, E. Carruthers, A. Curry, M. Kuroda, E. Fallows, J. Thomas, D. Sherman and M. Muldoon, Int. J. Food Microbiol., 2015, 198, 19–27 CrossRef CAS PubMed.
- S. Efrima and L. Zeiri, J. Raman Spectrosc., 2009, 40, 277–288 CrossRef CAS.
- M. Knauer, N. Ivleva, R. Niessner and C. Haisch, Anal. Sci., 2010, 26, 761–766 CrossRef CAS PubMed.
- H. Zhou, D. Yang, N. P. Ivleva, N. E. Mircescu, R. Niessner and C. Haisch, Anal. Chem., 2014, 86, 1525–1533 CrossRef CAS PubMed.
- W. R. Premasiri, D. T. Moir, M. S. Klempner, N. Krieger, G. Jones and L. D. Ziegler, J. Phys. Chem. B, 2005, 109, 312–320 CrossRef CAS PubMed.
- D. Weller, A. Andrus, M. Wiedmann and H. C. den Bakker, Int. J. Syst. Evol. Microbiol., 2015, 65, 286–292 CrossRef CAS PubMed.
- M. Mendonça, N. L. Conrad, F. R. Conceição, A. N. Moreira, W. P. da Silva, J. A. Aleixo and A. K. Bhunia, BMC Microbiol., 2012, 12, 275 CrossRef PubMed.
- Y. Liu, K. Chao, X. Nou and Y.-R. Chen, Sens. Instrum. Food Qual. Saf., 2008, 3, 100–107 CrossRef.
- J. Chen, B. Shen, G. Qin, X. Hu, L. Qian, Z. Wang, S. Li, Y. Ren and L. Zuo, J. Phys. Chem. C, 2012, 116, 3320–3328 CAS.
- A. J. Chung, Y. S. Huh and D. Erickson, Nanoscale, 2011, 3, 2903–2908 RSC.
- U. Huebner, K. Weber, D. Cialla, R. Haehle, H. Schneidewind, M. Zeisberger, R. Mattheis, H.-G. Meyer and J. Popp, Microelectron. Eng., 2012, 98, 444–447 CrossRef CAS.
- B. C. Galarreta, P. R. Norton and F. Lagugné-Labarthet, Langmuir, 2011, 27, 1494–1498 CrossRef CAS PubMed.
- C. A. Smyth, I. Mirza, J. G. Lunney and E. M. McCabe, Appl. Surf. Sci., 2013, 264, 31–35 CrossRef CAS.
- M. Suzuki, Y. Niidome, N. Terasaki, K. Inoue, Y. Kuwahara and S. Yamada, Jpn. J. Appl. Phys., 2004, 43, L554–L556 CrossRef CAS.
- F. C. Cabrera, P. H. B. Aoki, R. F. Aroca, C. J. L. Constantino, D. S. dos Santos and A. E. Job, J. Raman Spectrosc., 2012, 43, 474–477 CrossRef CAS.
- E. P. Hoppmann, W. W. Yu and I. M. White, IEEE J. Sel. Top. Quantum Electron., 2014, 20, 195–204 CrossRef.
- S. Uusitalo, J. Hiltunen, P. Karioja, S. Siitonen, V. Kontturi, R. Myllyla, M. Kinnunen and I. Meglinski, J. Eur. Opt. Soc, Rapid Publ., 2015, 10, 15043 CrossRef.
- S. Z. Oo, R. Y. Chen, S. Siitonen, V. Kontturi, D. A. Eustace, J. Tuominen, S. Aikio and M. D. B. Charlton, Opt. Express, 2013, 21, 18484–18491 CrossRef CAS PubMed.
- K. Maximova, A. Aristov, M. Sentis and A. Kabashin, Nanotechnology, 2015, 26, 065601 CrossRef CAS PubMed.
- S. Besner, A. Kabashin and M. Meunier, Appl. Phys. A, 2007, 88, 269–272 CrossRef CAS.
- M. Kögler, B. Zhang, L. Cui, Y. Shi, M. Yliperttula, T. Laaksonen, T. Viitala and K. Zhang, Sens. Actuators, B, 2016, 230, 411–421 CrossRef.
- T. Rojalin, L. Kurki and T. Laaksonen, Anal. Bioanal. Chem., 2016, 408, 761–774 CrossRef CAS PubMed.
- Z. Tian, B. Ren, J. Li and Z. Yang, Chem. Commun., 2007, 34, 3514–3534 RSC.
- P. Blandin and K. Maximova, J. Mater. Chem. B, 2013, 1, 2489–2495 RSC.
- U. Tamer, İ. H. Boyacı, E. Temur, A. Zengin, İ. Dincer and Y. Elerman, J. Nanopart. Res., 2011, 13, 3167–3176 CrossRef CAS.
- H. Butler, S. Fogarty and J. Kerns, Analyst, 2015, 140, 3090–3097 RSC.
- T. Szymborski, E. Witkowska and W. Adamkiewicz, Analyst, 2014, 139, 5061–5064 RSC.
- M. Çulha, M. M. Yazıcı, M. Kahraman, F. Şahin and S. Kocagöz, J. Nanotechnol., 2012, 297560 Search PubMed.
- B. Luo and M. Lin, J. Rapid Methods Autom. Microbiol., 2008, 16, 238–255 CrossRef CAS.
- S. Vohník, C. Hanson, R. Tuma, J. A. Fuchs, C. Woodward and G. J. Thomas, Protein Sci., 1998, 7, 193–200 CrossRef PubMed.
- C. Fan, Z. Hu, A. Mustapha and M. Lin, Appl. Microbiol. Biotechnol., 2011, 92, 1053–1061 CrossRef CAS PubMed.
- H. Al-Qadiri and M. Lin, J. Food Sci., 2008, 73, M54–M61 CrossRef CAS PubMed.
- K. Maquelin, C. Kirschner, L.-P. Choo-Smith, N. van den Braak, H. P. Endtz, D. Naumann and G. Puppels, J. Microbiol. Methods, 2002, 51, 255–271 CrossRef CAS PubMed.
- L. Cui, P. Chen, B. Zhang, D. Zhang and J. Li, Water Res., 2015, 87, 282–291 CrossRef CAS PubMed.
- L. Chen and N. Mungroo, J. Nanobiotechnol., 2015, 13, 45 CrossRef PubMed.
- Y. Liu, Y. Chen, X. Nou and K. Chao, Appl. Spectrosc., 2007, 61, 824–831 CrossRef CAS PubMed.
- X. Lu, H. Al-Qadiri, M. Lin and B. Rasco, Food Bioprocess Technol., 2011, 4, 919–935 CrossRef.
- K. Maquelin and L. Choo-Smith, Anal. Chem., 2000, 72, 12–19 CrossRef CAS PubMed.
- M. Harz, P. Rösch and J. Popp, Cytometry, Part A, 2009, 75, 104–113 CrossRef CAS PubMed.
- G. Long and J. Winefordner, Anal. Chem., 1983, 55, 712A–724A CrossRef CAS.
- N. Gnanou Besse, N. Audinet, A. Kérouanton, P. Colin and M. Kalmokoff, Int. J. Food Microbiol., 2005, 104, 123–134 CrossRef PubMed.
Footnote |
| † S. Uusitalo and M. Kögler contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2016 |