Interaction between carisoprodol and bovine serum albumin and effect of β-cyclodextrin on binding: insights from molecular docking and spectroscopic techniques†
Abstract
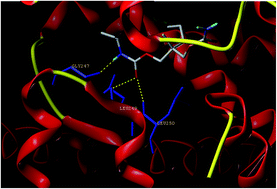
Biomolecular interactions of carisoprodol (CAP) with bovine serum albumin (BSA) have been studied by fluorescence and UV-visible spectroscopy and confirmed by multispectroscopic methods including molecular docking studies. The intrinsic intensity of BSA was quenched by a dynamic quenching mechanism. The binding constant and number of binding sites were calculated according to the Stern–Volmer equation. The effect of β-cyclodextrin on the binding has been studied. Thermodynamic parameters were calculated which reveal the involvement of hydrophobic interactions in the binding. Based on Förster's theory of non-radiation energy transfer, the average binding distance (r) between BSA and CAP was evaluated. Spectral results showed that the binding of CAP to BSA induced conformational changes in BSA. A molecular docking study confirmed the drug binding sites and interaction of CAP with amino acid residues.

 Please wait while we load your content...
Please wait while we load your content...