Rapid amplification of Mycobacterium tuberculosis DNA on a paper substrate†
Prasad Shetty,
Dipayan Ghosh,
Minakshi Singh,
Aparna Tripathi and
Debjani Paul*
Department of Biosciences and Bioengineering, Indian Institute of Technology Bombay, Powai, Mumbai 400076, India. E-mail: debjani.paul@iitb.ac.in
First published on 6th June 2016
Abstract
We have amplified an 84 bp fragment from the insertion sequence IS6110 of Mycobacterium tuberculosis (MTB) DNA on a paper substrate in 10 min. Tuberculosis (TB) is a highly infectious disease with a high mortality rate in the developing countries. Cheap and rapid screening assays are needed at the point of care for timely intervention and treatment. While PCR-based detection of TB is faster than bacterial culture and more specific compared to microscopy, PCR is not viable as a routine screening test in many low-income countries where TB is endemic due to the requirement of an expensive thermocycler. Disease screening based on isothermal amplification techniques is, therefore, being explored as an alternative to PCR. Successful isothermal amplification of DNA from HIV, H1N1, Chlamydia trachomatis, E. coli, etc. on cheap and disposable paper substrates has been reported in the literature. Isothermal amplification of MTB DNA on paper has been challenging due to the high GC content (65%) of MTB genome. Here we report helicase dependent amplification of a fragment of MTB DNA on a paper substrate in 10 min starting from 100 copies of the template using inexpensive heat sources, such as hot plates and hand warmers. The enzyme mix used to amplify DNA can be spotted and stored dry on paper at ambient temperatures for more than a month. The DNA amplified on paper can be detected by incorporating a suitable fluorescence marker in the reaction mixture or by directly loading the paper in a standard gel electrophoresis set up. Finally, as a surrogate to real clinical sputum samples, MTB DNA was successfully amplified on paper in the viscous environment of artificial sputum.
Introduction
Tuberculosis (TB), a highly contagious disease caused by the pathogen Mycobacterium tuberculosis (MTB), was declared a global emergency by the WHO in 1993.1 In 2014 ∼9.6 million people newly contracted TB and ∼1.5 million died due to a lack of timely diagnosis and available treatment options. Out of the 9.6 million new TB cases, India alone accounted for ∼2–2.3 million.2 Among the current diagnostic approaches, smear microscopy is widely used in low resource settings to identify mycobacteria. But a positive smear test fails to distinguish between MTB and non-tuberculous mycobacteria (NTM). While mycobacterial culture remains a gold standard for TB diagnosis, it takes several days or weeks to confirm the presence of MTB. TB detection techniques based on serological tests are faster, but they have much lower sensitivity compared to culture.3 Nucleic acid amplification techniques, e.g. PCR, are highly sensitive, specific and rapid (taking only a few hours from sample to answer). The most widely used commercially available PCR-based tuberculosis detection platform is the GeneXpert (Cepheid).4 Genedrive5 and COBAS (Roche)6 are other PCR-based assays available for TB detection. These systems are typically available in well-equipped tertiary hospitals. None of these platforms are targeted to cater to the bottom of the healthcare pyramid (i.e. the primary healthcare centres in rural areas) due to their relatively high running costs, requirement of highly trained personnel and the need for continuous line power. The subsidized cost of GeneXpert platform is USD 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–17
000–17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, while the yearly calibration cost of the system is USD 1800. The cost per test is USD 9.98, which is beyond the reach of a large part of the affected population, given the prolonged nature of the treatment and the need for regular monitoring. According to WHO, there is a strong and unmet need for a “rapid, sputum-based, molecular test for microscopy centres” for use in countries with a heavy tuberculosis burden.3 Therefore, our goal was to demonstrate specific amplification of MTB DNA on a paper substrate as a first step towards an affordable, rapid, disposable, ‘yes/no’ type molecular test for TB that can be used for pre-screening at the point-of-care. Such a test would be useful for identifying those patients who need to undergo further tests using more powerful platforms such as, the GeneXpert.
500, while the yearly calibration cost of the system is USD 1800. The cost per test is USD 9.98, which is beyond the reach of a large part of the affected population, given the prolonged nature of the treatment and the need for regular monitoring. According to WHO, there is a strong and unmet need for a “rapid, sputum-based, molecular test for microscopy centres” for use in countries with a heavy tuberculosis burden.3 Therefore, our goal was to demonstrate specific amplification of MTB DNA on a paper substrate as a first step towards an affordable, rapid, disposable, ‘yes/no’ type molecular test for TB that can be used for pre-screening at the point-of-care. Such a test would be useful for identifying those patients who need to undergo further tests using more powerful platforms such as, the GeneXpert.
Isothermal enzymatic amplification of DNA requires a single incubation temperature and can be performed without a thermocycler. Several isothermal amplification techniques have been reported in the past, such as, loop-mediated amplification (LAMP), recombinase polymerase amplification (RPA), helicase-dependent amplification (HDA), nucleic acid sequence based amplification (NASBA), rolling circle amplification (RCA), strand displacement amplification (SDA), etc.7 There are a few commercially available MTB diagnostic kits that employ isothermal amplification, e.g. LoopAMP kit (Eiken, Japan) relying on LAMP8 and EasyNAT kit (Ustar, China) based on cross-priming amplification (CPA).9 Each of these amplification schemes requires four or more sets of primers. HDA utilizes the helicase enzyme instead of heat to unwind double-stranded DNA at 65 °C and requires only one set of primers. HDA can amplify genomic DNA from crude samples, such as, blood10 and bacterial cells,11 which is an important requirement for point-of-care diagnostic assays. Therefore, we chose to work with HDA to develop our TB screening assay.
Paper is cheap, abundant, portable, disposable and can wick samples by capillary action without using any external pump. As a result, paper has been widely used for diagnostic applications, particularly for separation and analysis of proteins and small molecules.12–14 Paper-based devices for nucleic acid amplification have been reported in the literature.11,15–22 In some reports paper was used only to store nucleic acids, while the amplification took place in solution.11,17–22 In another technique, primers were immobilized on paper and solid phase amplification was carried out.22 This method needs an additional primer immobilisation step. Recently Rohrman et al.15 successfully amplified HIV DNA on a paper/plastic microfluidic chip, while Linnes and others11 amplified Chlamydia trachomatis DNA on a paper substrate placed inside a pipette tip. The same group has recently reported extraction and amplification of RNA from Influenza A (H1N1) on a polyethersulfone matrix.16 Connelly et al. have integrated sample preparation, LAMP based amplification and fluorescent detection of Escherichia coli on a paper machine platform.17
There are currently no reports on successful amplification of MTB DNA on a paper substrate, which is the most crucial step towards the development of a paperfluidic molecular test for TB screening. Isothermal amplification of MTB DNA at a lower temperature has been challenging as the MTB genome has a relatively higher (65.6%) GC content.23 Higher GC content increases the DNA melting temperature, leads to secondary structure formation, etc.24 Unlike PCR, isothermal amplification does not have a heat denaturation step. Enzymatic unwinding of DNA becomes more challenging when the GC content is high. Therefore, the recommended conditions for HDA are that (i) the GC content of the primers should not exceed 60%, and (ii) the GC content should preferably be similar to the GC content of the template DNA.25 So, the regions of the MTB genome that can be amplified by HDA are those where the GC content is less than 60%. We report here for the first time rapid (∼10 min) amplification of an 84 bp fragment from the insertion sequence 6110 (IS6110) of MTB on a paper substrate. The IS6110 sequence was chosen because it is conserved in M. tuberculosis complex.26 We used HDA to amplify MTB DNA in a reaction volume as low as ∼4 μL. We also showed that the reagents for HDA could be stored dry on paper at room temperature in a stable form. Our optimized assay can amplify and detect as little as 100 copies of the template on a paper substrate. Finally, we have successfully performed HDA of MTB DNA in the presence of viscous artificial sputum in an attempt to mimic clinical sputum samples. Our novel paper-based MTB amplification assay is a crucial step towards developing a molecular pre-screening test for tuberculosis.
Materials and methods
Equipment and chemicals
We used chromatography paper (Whatman grade 1, Mumbai, India) as the substrate for all paper-based HDA reactions. A heat sealer (Impulse sealer, Model: PFS-300P, 450 W) was used to heat seal the plastic pouch containing the paper substrate prior to HDA. Temperature measurement was done by means of an IR thermometer (IRX-63, HTC Instruments, India). A hot plate (IKA C-MAG HS 7, Cole Parmer, India), a thermal cycler (MJ mini, Bio-Rad, India) and a pair of pocket hand warmers (Aldi, UK) were used as heating sources. IsoAmp III enzyme mix for HDA was purchased from Biohelix Inc. (Beverly, USA). Filter tips (ART barrier specialty pipette tips), deoxyribonucleotide triphosphate mix (dNTP), deoxyadenosine triphosphate (dATP) and DNA ladder (O'GeneRuler ultra low range 10–300 bp ladder) were purchased from Thermo Scientific (Mumbai, India). We got the amplification primers synthesized by Sigma Aldrich (Bangalore, India). DAPI, methylcellulose and egg yolk emulsion were also from Sigma Aldrich (Mumbai, India). Ethidium bromide was bought from Merck Millipore (Mumbai, india). PicoGreen (Quant-iT PicoGreen dsDNA assay kit) and SYBR Green 1 (nucleic acid gel stain) were purchased from Life Technologies (Mumbai, India). Propidium iodide was obtained from HiMedia (Mumbai, India). SeaKem LE Agarose was purchased from Lonza (Mumbai, India). The electrophoresis unit was from Genetix (Mumbai, India). PerkinElmer Geliance 1000 imaging system was used for imaging the agarose gels. The concentration of DNA was measured using a NanoDrop instrument (NanoPhotometer® P 300, IMPLEN). Grey scale images of fluorescent signals were captured using an inverted fluorescence microscope (Nikon eclipse Ti-E) and analysed using the free image processing software ImageJ (NIH).HDA reaction composition
An 84 bp fragment from the insertion sequence 6110 (IS6110) of MTB genomic DNA (H37Rv strain) was first amplified by PCR and the purified PCR product was used as the amplification template for HDA on paper. For performing solution phase HDA (positive control reactions), either MTB genomic DNA or the PCR-amplified template was used with a previously reported26 set of primers. HDA reaction parameters (incubation temperature, time, reaction volume, etc.) were initially optimized in solution and then transposed to the paper substrate. The protocol recommended by Biohelix Inc. for a single-step HDA reaction involves incubation for 90 min at 65 °C. The other suggested parameters are product size ∼80–120 bp, primer size ∼24–33 bp, primer Tm ∼ 68–74 °C, product Tm ∼ 68–75 °C and primer GC content ∼35–60%. We used the commercially available IsoAmp III enzyme mix for HDA, which includes E. coli UvrD helicase, accessory protein MutL, phage T4 gene 32 protein (i.e. single-stranded DNA binding protein) and exo-Klenow fragment of DNA polymerase I. We used 1× concentration of the enzyme for solution phase reactions. The remaining reaction components were 1× annealing buffer II, 4 mM MgSO4, 40 mM NaCl, 0.4 mM of each dNTP, 4.65 mM additional dATP, forward and reverse primers (0.12 μM each), DNA template (typically 1010 copies of the PCR-amplified 84 bp MTB DNA) and DNase-free water. For DNA amplification in artificial sputum, 2% methylcellulose and 10% egg yolk emulsion was spiked with 108 to 1010 copies of an 84 bp MTB DNA.Transposing HDA to paper
Fig. 1 shows a schematic diagram of our paper-based reaction platform. A 5 mm diameter paper disc was the substrate for the HDA reaction. The paper disc was first treated at 75 °C for 15 min to inactivate any DNases present due to previous handling, then soaked in a sterile BSA solution (0.25%) overnight and dried in air. A 4 μL solution containing all amplification reagents except the template DNA was spotted on the BSA-treated paper. For all paper-based HDA reactions, the enzyme concentration was chosen to be three times higher than the recommended concentration for solution-based reactions. The paper was dried in air for 1 h and stored at room temperature until further use. Before each HDA reaction, the required amount of template DNA in 4 μL of DI water was added to wet the paper disc. The wet paper substrate was heat-sealed inside a polyethylene pouch (10 mm × 10 mm) for amplification. HDA was performed with alternative heat sources, such as, hot plate (65 °C) or a pair of hand warmers (56 °C) along with a thermocycler programmed at a constant temperature.Detection of the amplified DNA
After amplification, the pouch was cut open to take out the paper substrate containing amplified DNA and dried in an oven at 65 °C for 10 minutes. The piece of paper was then directly loaded into the well of a 4% agarose gel stained with ethidium bromide (1.27 μM final concentration). We also explored fluorescence detection of the amplified DNA where a fluorescent dye was directly added into the amplification mix. Ethidium bromide (EtBr, 1.27 μM), propidium iodide (PI, 500 nM), 4′,6-diamidino-2-phenylindole (DAPI, 300 nM), SYBR Green 1 (SG, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 final dilution) and PicoGreen (PG, 1
000 final dilution) and PicoGreen (PG, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 final dilution) were initially explored to find the most suitable dye to visualize DNA on paper. The fluorescence intensity after amplification was measured both with and without washing protocols to remove excess unbound dye. The paper disc was washed with DI water to remove any unbound fluorescent dye. During the wash step, the paper disc was first placed on top of a folded tissue paper and 10 μL of DI water was pipetted directly on it. The excess water was wicked by the tissue paper. The wet tissue was replaced by a fresh tissue and the process was repeated three times. After initial trials with different dyes, PG was chosen. All amplification reactions were done in triplicates with two negative controls, e.g. no enzyme control (HDA without any enzyme) and no template control (HDA without any template DNA). Fluorescence imaging was done using a UV gel doc for EtBr (ex/em: 300 and 360 nm/590 nm) and SG (ex/em: 290, 380 and 497 nm/520 nm). Nikon Eclipse Ti-U fluorescence microscope was used to image amplified DNA on paper discs spotted with DAPI (ex/em: 364 nm/454 nm), PI (ex/em: 535 nm/617 nm) and PG (ex/em: 480 nm/520 nm). Grey scale images acquired by the microscope were analysed by ImageJ to obtain the fluorescence intensities.
400 final dilution) were initially explored to find the most suitable dye to visualize DNA on paper. The fluorescence intensity after amplification was measured both with and without washing protocols to remove excess unbound dye. The paper disc was washed with DI water to remove any unbound fluorescent dye. During the wash step, the paper disc was first placed on top of a folded tissue paper and 10 μL of DI water was pipetted directly on it. The excess water was wicked by the tissue paper. The wet tissue was replaced by a fresh tissue and the process was repeated three times. After initial trials with different dyes, PG was chosen. All amplification reactions were done in triplicates with two negative controls, e.g. no enzyme control (HDA without any enzyme) and no template control (HDA without any template DNA). Fluorescence imaging was done using a UV gel doc for EtBr (ex/em: 300 and 360 nm/590 nm) and SG (ex/em: 290, 380 and 497 nm/520 nm). Nikon Eclipse Ti-U fluorescence microscope was used to image amplified DNA on paper discs spotted with DAPI (ex/em: 364 nm/454 nm), PI (ex/em: 535 nm/617 nm) and PG (ex/em: 480 nm/520 nm). Grey scale images acquired by the microscope were analysed by ImageJ to obtain the fluorescence intensities.
Results and discussion
Choice of the isothermal amplification technique
We opted for an isothermal amplification technique over PCR to eliminate the need for an expensive thermocycler in smaller district hospitals. Our choice of the isothermal amplification technique was guided by the following requirements. (1) The technique should work with a single set of primers, making the transition between PCR and HDA easier. (2) It should specifically amplify the target DNA at a single incubation temperature without any initial heat-assisted denaturation step. This is necessary to eliminate the requirement of heat sources capable of cycling between multiple temperatures. (3) The technique should be able to amplify DNA from raw samples such as, blood and sputum (keeping in mind the future development of the amplification platform). Among the existing techniques, LAMP,7 SDA7 and CPA27 were not suitable, as they require more than one set of primers. While NASBA, RPA, HDA, RCA, etc. can work with a single set of primers, NASBA amplifies RNA targets7 and RCA has a low tolerance to unprocessed biological samples.28 RPA works between 37 °C and 42 °C, but the commercially available RPA kit recommends sequential re-hydration and activation of the reaction mixture by addition of magnesium acetate for the reaction to start. On the other hand, all components of HDA can be added right at the start of the reaction and no further user intervention is needed. RPA also has low sensitivity when blood samples are used,29–31 but HDA can amplify DNA from raw specimens, such as, blood and bacterial cells.10,11 Since the ability to work with raw samples is important for point-of-care diagnostic assays, we chose HDA over RPA to develop our amplification platform.Sealing the substrate
The advantage of developing a paper-based reaction platform is that the entire reaction can potentially be printed on the paper substrate similar to a technique recently reported by our group.32 We chose chromatography paper because it would have minimal chemical additives to inhibit DNA amplification. Since the recommended protocol for HDA involves incubation at 65 °C for 90 min, it is essential to properly seal the paper substrate to prevent evaporation of the reaction mixture. We started by exploring heat-resistant Scotch and polyimide tapes for sealing. However, the adhesive material present on these tapes inhibited DNA amplification (data not shown). The pores of the paper substrate were blocked when we laminated the paper.Therefore, in all our subsequent experiments, the paper substrate was put in a 10 mm × 10 mm polyethylene pouch and sealed using a rapid heat sealer. Two polyethylene (PE) sheets were cut to 10 mm × 10 mm dimension. These sheets were cleaned with 70% ethanol and DNase-free water before use to remove any impurities present. The PE sheets were further incubated at 75 °C for 15 min to inactivate any DNases present due to previous handling. Since PE is a thermoplastic, both the sheets can be sealed together using a heat sealer. The PE sheets were first sealed along three sides leaving one end unsealed for introduction of the paper disc. The paper disc with dried reagents was put into the PE pouch using sterilized tweezers and DNA sample was pipetted onto it. Then the open end of the pouch was sealed using the heat sealer. While sealing, we ensured that the paper disc was well inside the pouch and at least 3–4 mm away from the heat sealing region. The heat sealing requires ∼1 s to seal the pouch at 100 °C (the temperature was measured using an IR thermometer). We also checked the temperature of the reaction mix on paper during the sealing step and found that it was unaffected, most likely due to the low thermal conductivity of PE.
Substrate treatment prior to DNA amplification
Our initial attempts to amplify DNA on untreated chromatography paper were not successful due to possible non-specific adsorption of DNA and enzymes on the paper surface. To compensate for this loss, we used a higher (3×) concentration of enzymes. We also found that the amplification on paper fails unless the paper substrate is pre-treated with BSA or surfactants to further reduce non-specific adsorption. As expected, HDA reactions in solution did not suffer from this problem. Tween 20 (0.05%) and Triton X-100 (0.05%) were other possible options, as they did not inhibit HDA (Fig. S1 in ESI†).Dry storage of enzymes on paper
The enzymes used for amplifying DNA are typically stored at −20 °C or −80 °C to retain their activity over time. Cold chain transport or refrigerated storage facilities are not always available in the endemic regions of many developing countries. Use of these facilities adds to the final cost of the assay. Therefore, it is important to explore the stability of amplification reagents at room temperature. As a first step towards this goal, we spotted and air-dried the enzymes on the paper substrate at room temperature for an hour and revived it just prior to amplification. HDA was performed in regular intervals (data not shown) from day 1 to day 34. Fig. 2 shows the gel electrophoresis results of amplification performed with enzymes stored on paper for 34 days. Since ethidium bromide strongly binds to cellulose (even without any DNA), the paper inserted in the gel well also gives a bright fluorescence. We found that dry storage of the enzyme mix at ambient temperature (ranging from 25 °C to 40 °C) up to 34 days had no significantly deleterious effect on its amplification efficiency. This is possibly because the enzyme mix contains polyethylene glycol (PEG) and other high molecular weight sugar molecules.33 We did not explore dry storage of the HDA enzyme mix beyond 34 days.Use of cheaper heat sources
We chose to perform HDA instead of PCR to obviate the need for thermocyclers costing upwards of a few thousand USD. Therefore, we explored HDA with other heat sources, such as, a hot plate (set at 65 °C) and a pair of hand warmers (56 °C). Temperature-controlled ceramic top hot plates typically cost about USD 100, while a pair of hand warmers cost less than USD 1. Programme for Appropriate Technology in Health (PATH) developed an inexpensive and portable chemical heater for isothermal amplification in PCR tubes and demonstrated its use by amplifying HIV-1 DNA.34 The Klapperich group had demonstrated HDA of Clostridium difficile DNA in a cyclic olefin polymer chip using a toe warmer placed inside a ventilated Styrofoam cup.35 Since MTB DNA has a relatively higher GC content (65.6%) compared to C. difficile and HIV-1, it was not immediately obvious whether MTB DNA amplification could be performed on paper substrate at a fixed lower temperature using heat sources other than a thermocycler. While the recommended temperature for HDA using the Biohelix tHDA enzyme kit is 65 °C, we found that it can amplify MTB DNA even at 50 °C (Fig. S2, ESI†). No bands were seen for temperatures below 50 °C. Fig. 3 compares the amplification efficiency of HDA using three different heat sources, e.g. thermocycler (TC), hot plate (HP) and hand warmers (HW). The hot plate was set at 65 °C prior to HDA reaction. The paper device was stuck to the hot plate using cellotape to ensure uniform thermal contact. The pair of hand warmers maintained a temperature of 56 ± 0.5 °C for ∼90 min when kept inside a Styrofoam box. We sandwiched a paper device between the hand warmers for HDA. Control HDA reactions (in solution and on paper) were performed using a thermocycler set at 65 °C (recommended temperature) and 56 °C (temperature of a hand warmer) respectively. Our results showed that MTB DNA could be amplified on paper at temperatures as low as 50 °C. The band intensities obtained in reactions using thermocycler, hot plate and hand warmers were comparable, indicating that cheaper heat sources can be used to amplify MTB DNA for point-of-care testing.Minimizing the reaction volume and the reaction time
One of our goals was to determine the minimum sample volume needed for HDA reactions on paper. A smaller reaction volume would reduce the requirement of expensive reagents. The design of the paperfluidic chip in many of the reported amplification assays11,17–22 require the use of a larger reaction volume than what is actually necessary to wet the paper substrate. We estimated that the volume necessary to fully wet a 5 mm diameter chromatography paper disc is ∼4–5 μL. Since, the typical HDA reactions in solution use 50 μL reaction mix, we explored different reaction volumes (e.g. 50 μL, 20 μL, 10 μL and 4 μL) for paper-based reactions, while keeping the amount of template DNA constant. Reactions with the same volume in solution were used as positive controls. Fig. 4 shows the results of HDA with the same amount of template DNA (1010 copies), but with different reaction volumes. Some primer–dimer formation could be seen in the no template negative control. Since 84 bp bands in the gel could be seen with as little as 4 μL sample volume, all subsequent HDA reactions on paper were carried out with this volume, while varying the amount of template DNA as required.Next, we explored the minimum reaction time that would give a distinct band in the gel. Reaction times of 20 min, 15 min and 10 min were explored with 4 μL reactions containing 1010 copies of template DNA. We could see distinct gel bands after 10 min of HDA. The intensities of the bands for different reaction times were also comparable. Therefore, all further HDA reactions were performed with 4 μL volume and for 10 min. Any further reduction in HDA time was not explored, as the time taken for diagnosis beyond this point would typically be limited by sample processing and detection times.
Amplification in artificial sputum environment
Next, we amplified TB DNA in artificial sputum to check the efficacy of amplification on a paper substrate in the characteristic viscous environment of clinical sputum samples. Artificial sputum was used as a surrogate since the use of infected clinical sputum samples requires a biosafety level 3 facility. WHO has recommended a protocol for preparing artificial sputum with methylcellulose and egg emulsion.36 De Kantor et al.36 used 1% methylcellulose with emulsion from one egg, while Yamada and others37 used 2% methylcellulose and no other proteins. We combined both these protocols by adding 2% methylcellulose to 10% egg yolk emulsion. The proportion of methylcellulose was doubled to further increase the viscosity of the reaction mixture so that it mimics real samples better. The reaction mixture in artificial sputum was spotted on paper (in duplicates) and HDA was performed according to optimized protocols. A positive control (HDA reaction without artificial sputum) was also performed in solution. The results of HDA were determined by gel electrophoresis, as shown in Fig. 5. Our results show that the target band intensity is comparable to the band intensity of the positive control reaction. This result establishes that HDA can be used to successfully amplify MTB DNA in a viscous environment, typically mimicking the viscosity of certain clinical samples.Detection sensitivity
While we could amplify MTB DNA on paper in just 10 min, detecting the amplified DNA by gel electrophoresis added an extra 30–40 min to the total assay time. The detection sensitivity of the agarose gel was also lower compared to the DNA-binding dyes typically used in real-time detection. Therefore, we incorporated some common DNA-binding dyes (ethidium bromide, propidium iodide, DAPI, Sybr Green I and PicoGreen) into the HDA reaction mix and compared their fluorescence signals on paper.We had earlier noticed that chromatography paper strongly fluoresces in the UV light. When ethidium bromide without any DNA (a negative control) was added to paper, it also gave a bright fluorescence unlike the free dye in solution. Therefore, it was clearly not viable to distinguish the signal from the amplified DNA on paper using ethidium bromide as the intercalating dye. HDA on paper using propidium iodide, DAPI or Sybr Green I also did not increase the fluorescence intensity by any significant amount compared to the negative controls (i.e. no template and no enzyme controls). It should be noted that all of these dyes can be excited with UV (either as primary or secondary excitation). Paper itself gives a lot of autofluorescence in UV excitation. Adding a wash step before measuring the fluorescence intensities with these dyes did not improve the signal-to-noise ratio. Fig. S3 in the ESI† shows the results of the initial trials using different DNA-binding dyes. Unlike the other dyes, PicoGreen (PG) at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 final dilution did not give much background signal on paper. PG is excited using blue light (480 nm) and emits a green (520 nm) signal. It has very weak absorption in the UV region. The fluorescent intensity of HDA using PG was significantly higher compared to the negative controls even without a wash step. This is because PG is very sensitive with a detection range in picograms.38 The absence of a wash step prior to detection is particularly useful as integrating sequential steps with automated flow control on paperfluidic platforms can be quite challenging. Henceforth, all fluorescence detection experiments on paper were performed by adding PG in the HDA reaction mix.
400 final dilution did not give much background signal on paper. PG is excited using blue light (480 nm) and emits a green (520 nm) signal. It has very weak absorption in the UV region. The fluorescent intensity of HDA using PG was significantly higher compared to the negative controls even without a wash step. This is because PG is very sensitive with a detection range in picograms.38 The absence of a wash step prior to detection is particularly useful as integrating sequential steps with automated flow control on paperfluidic platforms can be quite challenging. Henceforth, all fluorescence detection experiments on paper were performed by adding PG in the HDA reaction mix.
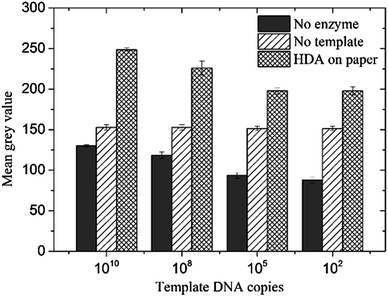
Early stage clinical samples may contain very little pathogenic DNA, and hence, it was important to estimate the sensitivity of our amplification platform. We varied the copy number of the template DNA from 1010 to 102 copies for 10 min HDA on paper. Fig. 6 shows a plot of the fluorescence intensity (mean grey value from three HDA reactions) vs. the template copy number. For all copy numbers, a 30–60% increase can be seen compared to the mean grey intensities of the two negative controls (no-template and no-enzyme reactions respectively). The slightly decreasing fluorescence intensity of the no-enzyme reaction with copy number was due to the presence of different amounts of template DNA. As expected, the intensities of no-template reactions are independent of the copy number. The lowest detection limit in our study is 100 copies of template DNA after 10 minutes of HDA. At this point, our assay can be used as a qualitative ‘yes/no’ type of detection test. Our future work will involve further optimization of the assay to develop it into a quantitative technique to estimate the number of pathogens.
In a separate experiment, we also verified the specificity of the MTB primers chosen for HDA by testing against another organism (M. smegmatis) from the Mycobacteria species. The primers did not amplify any sequence from M. smegmatis, while they worked fine with MTB (ESI, Fig. S4†).
Conclusions and future work
To the best of our knowledge, this is the first report of amplification of MTB DNA on a paper substrate. We have successfully amplified an 84 bp fragment of the MTB genome on a paper substrate in 10 min and with as little as 100 copies of the template DNA. The fuss-free design of our paperfluidic chip allows us to work with a small reaction volume (i.e. <5 μL) to reduce the use of expensive reagents. Since we employ isothermal amplification, we do not require a thermocycler, but can work with a cheap heat source (e.g. a hot plate or a pair of hand warmers). All HDA reagents, except the template DNA, can be pre-loaded and stored in a dried form on a 5 mm diameter paper disc. We have found that the dried enzyme mix remains stable on paper for at least 34 days at ambient temperature. The amplification reaction also works well in a viscous artificial sputum environment, as a surrogate for real clinical sputum samples. The primers that target IS6110 are specific for MTB (H37Rv strain) and do not amplify M. smegmatis (mc2155 strain) DNA. Finally, we have identified a suitable DNA-binding dye (PicoGreen) that can be incorporated into the amplification mix to detect the amplified DNA on paper. Use of this dye obviates any washing step, thus minimizing user intervention in the assay.A complete molecular TB diagnosis platform requires modules for sample lysis, DNA amplification and detection (visual/optical/electrochemical). Here we have demonstrated a proof of concept of isothermal amplification of MTB DNA on a paper substrate. Our TB amplification assay is rapid (∼10 min), minimally instrumented (using hand warmers or battery operated heaters), capable of storing HDA enzymes in a dried form at room temperature for more than a month and needs no user intervention after addition of sample. The PicoGreen fluorescence intensity from the target reaction appears to decrease slightly with a decrease in the template copy number. More extensive studies will be needed to develop this method into a viable quantitative detection technique. Therefore, our future work will be two-fold: (a) development of a compatible on-paper lysis protocol for Mycobacterium tuberculosis, as lysis of Mycobacteria on a paper substrate is challenging due to the presence of thick mycolic acid layer in the cell walls, and (b) further development of the fluorescence detection assay to enable more quantitative detection. We will also build a portable fluorescence reader to carry out the detection in resource-limited settings.
Acknowledgements
The authors acknowledge funding from the Innovative Young Biotechnologist Award (IYBA), Dept. of Biotechnology, Govt. of India, seed grant from IIT Bombay and Wadhwani Centre for Bioengineering, IIT Bombay. We thank Hinduja Hospital, Mumbai, and the Foundation for Medical Research, Mumbai, for providing MTB genomic DNA. We thank Sarika Mehra for providing M. smegmatis (mc2155) strain. The authors also thank Swati Patankar, Kiran Kondabagil, Anirvan Chatterjee, Kayzad Nilgiriwala and Kanchan Abjani for helpful discussions and feedback on the manuscript. Claudy D'Costa, Ammar Jagirdar and Nimisha Roy provided technical help.References
- TB: A global emergency, WHO report on the TB epidemic, 1994 Search PubMed.
- Global Tuberculosis report, World Health Organization, 2014 Search PubMed.
- UNITAID, Tuberculosis: Diagnostic Technology and Market Landscape, 3rd edn, 2014 Search PubMed
.
- Tuberculosis Diagnostics Xpert MTB/RIF Test, World Health Organisation, 2013 Search PubMed.
- P. Castan, A. de Pablo, N. Fernández-Romero, J. M. Rubio, B. D. Cobb, J. Mingorance and C. Toro, J. Clin. Microbiol., 2014, 52, 502–507 CrossRef PubMed
.
- G. V. Bloemberg, A. Voit, C. Ritter, V. Deggim and E. C. Böttger, J. Clin. Microbiol., 2013, 51, 2112–2117 CrossRef CAS PubMed
.
- L. Yan, J. Zhou, Y. Zheng, A. S. Gamson, B. T. Roembke, S. Nakayama and H. O. Sintim, Mol. BioSyst., 2014, 10, 970–1003 RSC
.
- S. Mitarai, M. Okumura, E. Toyota, T. Yoshiyama, A. Aono, A. Sejimo, Y. Azuma, K. Sugahara, T. Nagasawa, N. Nagayama, A. Yamane, R. Yano, H. Kokuto, K. Morimoto, M. Ueyama, M. Kubota, R. Yi, H. Ogata, S. Kudoh and T. Mori, Int. J. Tuberc. Lung Dis., 2011, 15, 1211–1217 CrossRef CAS PubMed
.
- R. McNerney, J. Cunningham, P. Hepple and A. Zumla, Int. J. Infect. Dis., 2015, 32, 81–86 CrossRef PubMed
.
- M. Vincent, Y. Xu and H. Kong, EMBO Rep., 2004, 5, 795–800 CrossRef CAS PubMed
.
- J. C. Linnes, A. Fan, N. M. Rodriguez, B. Lemieux, H. Kong and C. M. Klapperich, RSC Adv., 2014, 4, 42245–42251 RSC
.
- A. W. Martinez, S. T. Phillips and G. M. Whitesides, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 19606–19611 CrossRef CAS PubMed
.
- C. M. Cheng, A. W. Martinez, J. Gong, C. R. Mace, S. T. Phillips, E. Carrilho, K. A. Mirica and G. M. Whitesides, Angew. Chem., 2010, 122, 4881–4884 CrossRef
.
- G. E. Fridley, H. Le and P. Yager, Anal. Chem., 2014, 86, 6447–6453 CrossRef CAS PubMed
.
- B. Rohrman and R. R. Richards-Kortum, Lab Chip, 2012, 12, 3082–3088 RSC
.
- N. M. Rodriguez, J. C. Linnes, A. Fan, C. K. Ellenson, N. R. Pollock and C. M. Klapperich, Anal. Chem., 2015, 87, 7872–7879 CrossRef CAS PubMed
.
- J. T. Connelly, J. P. Rolland and G. M. Whitesides, Anal. Chem., 2015, 87, 7595–7601 CrossRef CAS PubMed
.
- USPTO, Molecular diagnostics amplification system and methods, US 8883487 B2 (Grant), 2014
.
- WIPO, Solid matrix for one step nucleic acid amplification, WO 2014041093 A1 (Application), 2014
.
- USPTO, Methods and apparatus for point-of-care nucleic acid amplification and detection, US 20120264116 A1 (Application), 2012
.
- JPO, Simple method for amplifying feces-derived gene, JP2006166711 (Application), 2006
.
- WIPO, Rapid test including genetic sequence probe, WO 2009128960 A2 (Application), 2009
.
- S. T. Cole, R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead and B. G. Barrell, Nature, 1998, 393, 537–544 CrossRef CAS PubMed
.
- A. Kumar and J. Kaur, Mol. Biol. Int., 2014, 937308, DOI:10.1155/2014/937308
.
- Thermophilic Helicase Dependent Amplification (tHDA) manual, http://www.biohelix.com/pdf/H0110S_full_version_BH_NG_eDatacard.pdf, accessed May 19 2016 Search PubMed.
- A. Motré, R. Kong and Y. Li, J. Microbiol. Methods, 2011, 84, 343–345 CrossRef PubMed
.
- G. Xu, L. Hu, H. Zhong, H. Wang, S. Yusa, T. C. Weiss, P. J. Romaniuk, S. Pickerill and Q. You, Sci Rep., 2012, 2, 246 Search PubMed
.
- P. Gill and A. Ghaemi, Nucleosides, Nucleotides, and Nucleic Acids, 2008, vol. 27(3), pp. 224–243 Search PubMed
.
- A. Abd El Wahed, A. El-Deeb, M. El-Tholoth, H. Abd El Kader, A. Ahmed, S. Hassan, B. Hoffmann, B. Haas, M. A. Shalaby, F. T. Hufert and M. Weidmann, PLoS One, 2013, 8(8), e71642 CAS
.
- M. Euler, Y. Wang, P. Otto, H. Tomaso, R. Escudero, P. Anda, F. T. Hufert and M. Weidmann, J. Clin. Microbiol., 2012, 50, 2234–2238 CrossRef CAS PubMed
.
- B. Rohrman and R. Richards-Kortum, Anal. Chem., 2015, 87, 1963–1967 CrossRef CAS PubMed
.
- A. Jagirdar, P. Shetty, S. Satti, S. Garg and D. Paul, Anal. Methods, 2015, 7, 1293–1299 RSC
.
- WIPO, Helicase dependent amplification of nucleic acids, WO2004027025A3 (Application), 2003
.
- J. Singleton, J. L. Osborn, L. Lillis, K. Hawkins, D. Guelig, W. Price, R. Johns, K. Ebels, D. Boyle, B. Weigl and P. LaBarre, PLoS One, 2014, 9(11), e113693 Search PubMed
.
- S. Huang, J. Do, M. Mahalanabis, A. Fan, L. Zhao, L. Jepeal, S. K. Singh and C. M. Klapperich, PLoS One, 2013, 8(3), e60059 CAS
.
- I. N. de Kantor, S. J. Kim, T. Frieden, A. Laszlo, F. Luelmo, P.-Y. Norval, H. Rider, P. Valenzuela and K. Weyer, Laboratory services in tuberculosis control. Organization and management. Part I, WHO, Geneva, Switzerland, 1998, p. 44 Search PubMed
.
- H. Yamada, S. Mitarai, L. Aguiman, H. Matsumoto and A. Fujiki, Int. J. Tuberc. Lung Dis., 2006, 10(8), 899–905 CAS
.
- A. I. Dragan, J. R. Casas-Finet, E. S. Bishop, R. J. Strouse, M. A. Schenerman and C. D. Geddes, Biophys. J., 2010, 99, 3010–3019 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra07529k |
| This journal is © The Royal Society of Chemistry 2016 |