In silico approach towards lipase mediated chemoenzymatic synthesis of (S)-ranolazine, as an anti-anginal drug†
Ganesh Sawant,
Saptarshi Ghosh,
Sooram Banesh ,
Jayeeta Bhaumik and
Uttam Chand Banerjee*
,
Jayeeta Bhaumik and
Uttam Chand Banerjee*
Department of Pharmaceutical Technology (Biotechnology), National Institute of Pharmaceutical Education and Research, Sec-67, S. A. S. Nagar-160062, Punjab, India. E-mail: ucbanerjee@niper.ac.in
First published on 11th May 2016
Abstract
An in silico modelling based biocatalytic approach for the synthesis of drugs and drug intermediates in enantiopure forms is a rationalized methodology over the organo-chemical routes. In this study, enzyme-ligand based docking was carried out using (RS)-ranolazine, as the model drug for the screening of a suitable biocatalyst for the kinetic resolution of the racemic drug. The differential interaction of the two enantiomers with the lipase was analyzed on the basis of docking score and H-bond interaction with the amino acid residues, which helped to define the trans-esterification mechanism. Ranolazine [N-(2,6-dimethylphenyl)-2-[4-(2-hydroxy)-3-(2-methoxyphenoxy)propylpiperazin-1-yl]acetamide], an anti-anginal drug, significantly reduces the frequency of anginal attack and has also been used for the treatment of ventricular arrhythmias, and bradycardia. Various lipases were examined via computational as well as wet lab screening and Candida antartica lipase in the form of CLEA was the most efficient one for the (S)-selective kinetic resolution of (RS)-ranolazine, with highest conversion and enantiomeric excess. This is the first report of the chemo-enzymatic synthesis of (S)-ranolazine where the whole drug molecule was used for lipase catalysis. The present study showed that the combination of in silico studies and a classical wet lab approach could change the paradigm of biocatalysis.
1. Introduction
A computational approach for the exploration of suitable biocatalyst candidates for the enantioselective synthesis of drugs and drug intermediates is an emerging field in chemoenzymatic synthesis.1 Docking of small molecules with the enzymes/receptors reveals information about the binding nature and interactions with the enzyme residues. In chiral compound synthesis, enantioselectivity of the biocatalyst (enzyme) is a crucial factor and screening of catalysts in terms of selectivity is also a major concern.2 The combination of enzyme kinetics and molecular modelling was used to construct a model of how the enantiomers of secondary alcohols bind in the active site of this enzyme during transesterification reactions.3 The two enantiomers of a secondary alcohol are shown to bind in two different orientations in the active site. The enantioselectivity is caused by the difference in the free energy gaps between the ground states of the enantiomers and the corresponding transition states, which the enantiomers form with the enzyme.The thermodynamic ground states of the enantiomers are the same. Therefore, the enantioselectivity is determined by the difference in free energy between the two transition states.4 For small molecule docking, various softwares are available such as GOLD, MOE, Molegro Virtual Docking, Maestro Glide and each one of them has different scoring algorithm.5 According to various reports, biocatalysts are used in most of the biotransformation reactions with high regio- and stereoselectivity.6–9 Enzymes are environment friendly alternatives over conventional chemical catalysts as they can operate under ambient temperature and pressure.10,11 In pharmaceutical sector, the new biocatalytic approaches for the synthesis of drugs and drug intermediates are particularly appealing due to low cost and minimal use of toxic chemicals.8,12–14 Lipases belonging to the class of serine hydrolases (α/β hydrolases super family) can act on the organic–aqueous interface and their industrial potential is also well known.15–17 Notably, the mechanism of transesterification reaction of lipase through catalytic triad has been already described by various researchers.18,19 Incorporation of in silico studies along with the conventional biocatalytic approach reduces both time and production expenses significantly. It will help to select suitable enzyme preparations prior to the wet laboratory screening and importantly will give access to determine the mechanism of biocatalysis.
Ranolazine has been approved by US FDA in 2006 (ref. 20) for the treatment of chronic angina as it significantly reduces frequent anginal attack and replaces the use of short acting nitroglycerine.21 According to the report of the American Heart Association, cardiovascular disease is the leading cause of global mortality, accounting for 17.3 million deaths per year, which is expected to reach to more than 23.6 million by 2030.22 In this study, ranolazine [N-(2,6-dimethylphenyl)-2-[4-(2-hydroxy-3-(2-methoxyphenoxy)propyl)piperazin-1-yl]acetamide], member of the β blocker family used as an anti-anginal agent23 was selected as the model drug for the computational catalysis. Unlike other drugs used to treat chronic angina, ranolazine shows negligible effect on heart rate and blood pressure. The major mechanism is believed to be its inhibitory effect on late sodium influx during the cardiac potential of cardiomyocytes, which is particularly important in ischemic or hypoxic condition.24 By altering the intracellular sodium level, ranolazine can affect the sodium-dependent calcium channels during myocardial ischemia. Thus, ranolazine indirectly prevents the calcium overload and prevent cardiac ischemia.25 Of the two enantiomers, (S)-enantiomer showed the anti-anginal activity whereas the (R)-form showed anti-diabetic activity.26 The (S)-enantiomer is potentially stronger and better anti-arrhythmic drug than either the (R)-enantiomer or racemic ranolazine.26 Several chemical methods were reported for the synthesis of (RS)-ranolazine or enantiomers. A synthetic procedure continuing a single step was reported by Li et al. (2003) for the synthesis of (RS)-ranolazine with 96% yield.27 In another report, one step synthesis was described with 90% yield (Lu et al., 2004).28 An improved process for the synthesis of (RS)-ranolazine with 68% yield was reported by Aalla et al.29 These chemical synthetic methods involve costly catalysts and harsh reaction conditions, therefore they are neither cost-effective nor environment friendly. Biocatalysis can be the only alternative to make the process benign and economical. The first chemoenzymatic synthesis of both enantiomers of ranolazine was reported by Moen et al. (2005).30 A multistep synthetic procedure of ranolazine was also reported with 70% yield.31 There was a report on water based chemistry for the synthesis of (RS)/(R)/(S)-ranolazine by Kommi et al. (2013) with 93% yield.32 Regioselective ring opening by tangstate mediated catalysis for the synthesis of ranolazine was also reported by Pathare et al. (2013) with 85% yield.33
Here we report a retrosynthetic pathway for (S)-ranolazine as shown in Fig. 1. Lipase mediated kinetic resolution of the entire drug (also synthesized chemically) was carried out. This is the first instance where the complete drug has been used for the resolution. In silico approach has been used for the screening of lipases and to determine the enantioselective reaction mechanism. Overall, a combination of in silico and biocatalytic approach afforded the enantiopure ranolazine as a breakthrough in the treatment of cardiovascular diseases.
2. Results and discussion
2.1 Chemical synthesis of (RS)-ranolazine as a standard compound
Prior to optimizing and synthesizing the enantiopure ranolazine through biocatalytic route, it was necessary to prepare the standard compound via a chemical route. Recently a research group has developed a chemical route for the synthesis of enantiopure ranolazine. However, the major pitfall of the method was the use of expensive catalysts.32 We modified the chemical pathway to synthesize racemic ranolazine as described below.The first building block of ranolazine synthesis, 2-chloro-N-(2,6-dimethylphenyl)acetamide 2 was synthesized by the reaction of 2,6-dimethylaniline 1 and chloroacetylchloride with 97% yield (Scheme 1). Further, to incorporate the piperazine ring, compound 2 was treated with piperazine which resulted into the formation of product 3 with 70% yield (Scheme 1). To introduce the chiral center, N-(2,6-dimethylphenyl)-2-(piperazin-1-yl)acetamide 3 was treated with (RS)-epichloro-hydrin in the presence of SDS in water as a solvent which afforded product 4 with 70% yield (Scheme 1). This reaction is a good example of water based chemistry which makes the process green. Likewise, the racemic form of the drug molecule was synthesized by treating compound 4 with methoxyphenol in the presence of K2CO3 in water to afford (RS)-ranolazine 5 with 75% yield (Scheme 1). The use of water as a solvent, in the 3rd and 4th step of chemical synthesis makes it attractive in terms of industrial synthesis as it makes the process cost-effective and environment friendly. The current synthetic method is simpler than the previously reported one with comparable yield.32
2.2 Computational approach for the screening of biocatalysts
Being outstanding biocatalysts, enzymes play dominant role in most of the chemicals reactions.34 To understand the enzymatic reaction mechanism towards chemical reaction and screening of the suitable biocatalysts, in silico docking techniques play an important role.35 Computational screening of enzymes for biocatalytic reactions can open up a new avenue for green catalysis as it will provide a rational behind the study. Among a variety of docking tools available,36 the Molegro Molecular Viewer 2.5.0 (CLC Bio, Qiagen) program was used for the screening of lipase, as the biocatalyst towards the enantiopure synthesis of ranolazine. Docking of various lipases was carried out with (RS)-5 as the ligand (zinc 21984084) and molecular docking score and H-bond lengths between the ligand and the various amino acid residues of the enzymes were analyzed. The consequences showed varied docking scores, interaction with the amino acid residues of the typical enzyme and difference in H-bond lengths. Interestingly, the R and S forms of the substrate interacted differently with the same enzyme.1,37 Only specifically oriented substrate (R or S) can be interacted significantly with the amino acids present in active site of the enzyme due to diastereomeric product formation.In the present study, the interaction of OH group at the chiral centre of (RS)-5 with the amino acid and the distance of the H-bond was considered for the screening of the preeminent apposite enzyme for the kinetic resolution of (RS)-5. Out of 11 enzymes screened, the S form of (RS)-5 interacted significantly to the amino acid (Glu 67) of Candida antartica lipase A CLEA only, with H-bond length of 3.07 Å (Table 1). The Fig. 2 represents, –OH group of the ligand (S)-5, interacted with the carboxyl oxygen of C. antartica lipase A showing H-bond but in the opposite direction, here the –OH group of the ligand (R)-5 interacted with the –NH2 group of Phe 435 in C. antartica lipase A CLEA to form H-bond (3.12 Å), which is not an ideal interaction for the transesterification reaction (Fig. 3).
| Lipase | PDB IDa | (S)-Ranolazine | (R)-Ranolazine | ||||
|---|---|---|---|---|---|---|---|
| Docking scoreb | Steric interaction score | Interacting residues with H-bond distancec | Docking scoreb | Steric interaction score | Interacting residues with H-bond distancec | ||
| a Protein Data Bank identity number of the lipase.b Molecular docking simulation searches through numerous potential binding modes of the ligand (zinc 21984084) in the pocket denoted as the score, which is calculated by the following formula: score = Starget-ligand + Sligand.c H-bond distance of interacting amino acid residue of the enzyme and the ligand (ZINC 21984084). | |||||||
| Amano-30 | 2NW6 | −54.73 | −64.17 | No interaction | −67.14 | −76.35 | No interaction |
| Candida antartica lipase A CLEA | 3GUU | −57.28 | −59.02 | Asn63-3.034 Å; Glu67-3.071 Å | −40.67 | −46.18 | Phe435-3.12 Å |
| Lipase from C. rugosa 62316 | 1LPN | −36.0 | −44.63 | Arg183 2.465 Å; Asn222-2.113 Å | −33.94 | −41.61 | Gln187-3.05 Å |
| Lipase from C. rugosa 90860 | 1LPO | −36.19 | −35.89 | Asn222-3.119 Å; Trp221-2.652 Å | −40.56 | −45.6 | Asp40-3.170 Å |
| Lipase from C. rugosa L-1754 | 1GZ7 | −51.96 | −52.15 | Asn351-3.367 Å; Tyr299-2.659 Å | −47.13 | −50.41 | Tyr299-3.009 Å; Arg303-3.149 Å |
| Lipase from C. cylindricea | 1CLE | −60.34 | −65.74 | Ser209-2.526 Å | −57.98 | −62.22 | Ser209-2.515 Å |
| Lipase from Aspergillus niger | 1UZA | −47.56 | −53.62 | His113-3.282 Å; Tyr124-3.385 Å | −49.88 | −59.29 | No interaction |
| Lipase from P. cepacia | 3LIP | −48.69 | −50.27 | Asn239-3.106 Å; Thr231-2.728 Å | −51.46 | −58.23 | Leu234-3.106 Å; Ala226-1.730 Å |
| Porcine pancreatic lipase | 1PCN | −52.99 | −56.41 | Asn81-2.267 Å; Asn83-2.914 Å | −47.81 | −58.67 | Asn83-2.542 Å |
| Lipase from Mucor meihei | 3TGL | 297.39 | 280.62 | Ile193-3.133 Å | 226.7 | 204.99 | No interaction |
| C. antartica lipase in acrylic resin | 3W9B | −63.44 | −73.36 | No interaction | −66.71 | −77.76 | No interaction |
 | ||
| Fig. 2 Interaction of (S)-5 with CAL-A lipase CLEA. The interaction of amino acid residues of enzyme with the ligand was shown by dotted circle. | ||
 | ||
| Fig. 3 Interaction of (R)-5 with CAL-A lipase CLEA. The interaction of amino acid residues of enzyme with the ligand was shown by dotted circle. | ||
The docking score is more negative for (S)-5 [−57.28] than the other enantiomer (R)-5 [−40.67], which clearly indicates better interaction of the lipase with the (S)-form and selectivity for acylation.
This shows that how the interaction between the chiral substrate (ligand) and the lipase determines the enantioselectivity towards the synthesis of pure enantiomer. Among the other lipases, C. antartica lipase acrylic resin and Amano lipase have shown no significant interaction with the substrate (RS)-5 (Table 1) (Fig. 4 and 5).
 | ||
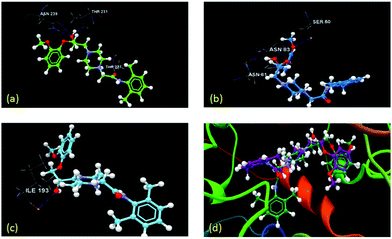
| Fig. 4 Docking result showing interaction of different lipases with (S)-5: (a) 2NW6, (b) 1LPN, (c) 1LPO, (d) 1GZ7, (e) 1CLE, (f) 1UZA. | ||
 | ||
| Fig. 5 Docking result showing interaction of different lipases with (S)-5: (a) 3LIP, (b) 1PCN, (c) 3TGL, (d) 3WNB. | ||
Docking with Mucor meihei lipase showed highly positive docking score, indicating poor binding and interaction with the substrate (Table 1). Most of the other lipases showed similar docking score for both the enantiomers, indicating the non-selective binding with the substrate (RS)-5 and resulting in non-selective acylation and low enantiomeric excess.
2.3 Screening of lipases for the kinetic resolution of (RS)-5
To support the in silico data, wet lab screening was performed with various commercial lipases to select the best enzyme on the basis of enantioselectivity, for the kinetic resolution of (RS)-5 (Table 2). Among the different sources, lipase from Candida cylindricea, C. rugosa CRL L1754, C. rugosa CRL 62316, C. rugosa CRL 90860, Aspergillus niger, immobilized lipase in sol–gel-AK from Pseudomonas cepacia, lipase acrylic resin from C. antartica lipase A, lipozyme of Mucor meihei lipase, cross linked enzyme aggregate of C. antartica lipase, Amano-30 lipase, and lipase from porcine pancreas were used for the screening. The biocatalytic reaction was carried out in toluene using vinyl acetate as the acyl donor (Scheme 2).| Name of lipase | eeSb (%) | eePc (%) | Conversiond (%) | Ee |
|---|---|---|---|---|
| a (RS)-5 (10 mM) in toluene (800 μL) was treated with vinyl acetate (200 μL, 2.5 mM) at 30 °C in presence of lipase (15 mg).b Enantiomeric excess of substrate calculated by using formula = (R − S)/(R + S) × 100.c Enantiomeric excess of product calculated by using formula = (S − R)/(S + R) × 100.d Conversions were calculated from enantiomeric excess of substrate (eeS) and product (eeP) using the formula: conversion (C) = eeS/(eeS + eeP).e E values were calculated using the formula: E = [ln(1 − C(1 + eeP))]/[ln(1 − C(1 − eeP))]. | ||||
| C. cylindricea lipase | 0.7 | 2.0 | 27 | 1.0 |
| C. antartica lipase A CLEA | 32 | 28 | 54 | 2.3 |
| Aspergillus niger lipase | 3.1 | 4.0 | 46 | 1.1 |
| C. rugosa 62316 | 1.0 | 5.5 | 15 | 1.1 |
| C. rugosa 90860 | 0.1 | 19 | 31 | 1.1 |
| C. rugosa L-1754 | 0.5 | 5.0 | 9.0 | 1.1 |
| Pseudomonas cepacia PCL | 5.2 | 2.3 | 69 | 1.0 |
| C. antartica acrylic resin | 2.3 | 4.0 | 39 | 1.1 |
| Amano-30 lipase | 5.2 | 2.3 | 69 | 1.5 |
| Porcine pancreatic lipase | 4.2 | 3.6 | 54 | 1.1 |
| Mucor meihei lipase | 0.7 | 1.0 | 42 | 1.1 |
Various lipases were screened and cross-linked enzyme aggregates (CLEA) of C. antartica lipase showed 53.55% conversion and enantioselectivity of substrate was very low (31.83%), which is better than the other lipases. To increase the enantiomeric excess of both substrate and product, optimization of various reaction parameters were carried out using CLEA of C. antartica lipase as the biocatalyst.
Lipase mediated biocatalytic reactions are highly affected by different physico–chemical parameters such as reaction medium, reaction time, temperature, enzyme and substrate concentration, solvent and type of acyl donor. The enantio-selectivity of the enzyme changes under different reaction conditions. To obtain the ideal reaction condition, effect of various reaction parameters were studied. Post optimization, the most suitable reaction condition was determined i.e. reaction time, 48 h; temperature, 30 °C; enzyme concentration, 30 mg mL−1; substrate concentration, 5 mM (Table 3).
| Physico–chemical parameters | Optimum value | eeSb (%) | eePc (%) | Conversiond (%) |
|---|---|---|---|---|
| a Kinetic resolution of (RS)-5 was carried out with Candida antartica lipase A CLEA.b Enantiomeric excess of substrate calculated by using formula = (R − S)/(R + S) × 100.c Enantiomeric excess of product calculated by using formula = (S − R)/(S + R) × 100.d Conversions were calculated from enantiomeric excess of substrate (eeS) and product (eeP) using the formula: conversion (C) = eeS/(eeS + eeP). | ||||
| Reaction time | 48 h | 73 | 100 | 42 |
| Temperature | 30 °C | 76 | 100 | 43 |
| Substrate concentration | 5 mM | 76 | 100 | 43 |
| Enzyme concentration | 30 mg mL−1 | 69 | 100 | 41 |
The effect of solvent on the kinetic resolution of (RS)-ranolazine 5 was studied by carrying out the biocatalytic reaction with solvents of different log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values (see ESI S2†). The result showed significant role of reaction medium on the enantioselectivity and conversion of the lipase. Amongst the six different solvents screened, diethyl ether was found to be ideal with conversion of 48% and both high eeS (90%) and eeP (98%). Also, the enantiomeric ratio (E) is significantly higher (323) for diethyl ether than the other solvents. Hence, further lipase mediated reactions were carried out using diethyl ether as the solvent (see ESI S3†). The role of acyl donor in the kinetic resolution of (RS)-5 was also studied and vinyl acetate (2.5 mM) was found to be the efficient one with 48% conversion (eeS 90%, eeP 99%). The enantiomeric ratio (E) was also higher (588) with vinyl acetate over other acyl donors used.
P values (see ESI S2†). The result showed significant role of reaction medium on the enantioselectivity and conversion of the lipase. Amongst the six different solvents screened, diethyl ether was found to be ideal with conversion of 48% and both high eeS (90%) and eeP (98%). Also, the enantiomeric ratio (E) is significantly higher (323) for diethyl ether than the other solvents. Hence, further lipase mediated reactions were carried out using diethyl ether as the solvent (see ESI S3†). The role of acyl donor in the kinetic resolution of (RS)-5 was also studied and vinyl acetate (2.5 mM) was found to be the efficient one with 48% conversion (eeS 90%, eeP 99%). The enantiomeric ratio (E) was also higher (588) with vinyl acetate over other acyl donors used.
2.4 Synthesis of (S)-ranolazine through deacylation
After successful synthesis of the enantiopure acylated derivative 6, we focused on the synthesis of our target drug i.e. enantiopure ranolazine. To synthesize (S)-ranolazine 5, the acylated derivative 6 was isolated after the biocatalytic reaction. The acylated derivative, (S)-6 was deacylated in aqueous K2CO3 at room temperature for 2 h (Scheme 3) which afforded (S)-ranolazine.3. Experimental
3.1 General experimental details
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2-propanol (9
2-propanol (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1); flow rate, 1 mL min−1 and column temperature of 25 °C.
1); flow rate, 1 mL min−1 and column temperature of 25 °C.3.2 Synthesis of 2-chloro-N-(2,6-dimethylphenyl)acetamide (2)
In the first step, 2,6-dimethylaniline 1 was treated with chloroacetyl chloride at 4 °C in dichloromethane to obtain 2-chloro-N-(2,6-dimethylphenyl)acetamide 2 with 97% practical yield. Recovered product was further analyzed by GC-MS and NMR spectroscopy. 1H NMR (CDCl3, 400 MHz) δ: 2.26 (s, 6H), 4.27 (s, 2H), 7.11–7.19 (m, 3H), 7.28 (s, 1H); 13C NMR (CDCl3, 100 MHz) δ: 18.32, 42.8, 127.92, 128.39, 132.67, 135.36, 164.38; m/z: 197.66 (M = C10H12ClNO).3.3 Synthesis of N-(2,6-dimethylphenyl)-2-(piperazin-1-yl)-acetamide (3)
For the second step, 2-chloro-N-(2,6-dimethylphenyl)-acetamide 2 was reacted with piperazine in methanol at 60 °C to obtain the product 3 with 70% practical yield. Recovered product was further analyzed by GC-MS and NMR spectroscopy. 1H NMR (CDCl3, 400 MHz) δ: 2.25 (s, 6H), 2.68–2.69 (d, J = 4 Hz, 4H), 2.97–2.99 (t, 4H), 3.18 (s, 2H), 7.10–7.12 (dd, J = 8 Hz, 3H), 8.67 (s, 1H); m/z: 247.95 (M = C14H21N3O).3.4 Synthesis of (RS)-2-(4-(3-chloro-2-hydroxyphenyl)piperazin-1-yl)-N-(2,6-dimethylphenyl)acetamide (4)
N-(2,6-Dimethylphenyl)-2-(piperazin-1-yl)acetamide 3 was treated with (RS)-epichlorohydrin in the presence of SDS using water as solvent to obtain (RS)-2-(4-(3-chloro-2-hydroxy-phenyl)piperazin-1-yl)-N-(2,6-dimethylphenyl)acetamide 4 with 70% isolated yield. Recovered product was analyzed by GC-MS and NMR spectroscopy. 1H NMR (CDCl3, 400 MHz) δ: 8.63 (s, 1H), 7.06–7.14 (m, 4H), 4.57 (s, 1H), 3.84–3.88 (dd, J = 16 Hz, 2H), 3.68–3.70 (d, J = 8 Hz, 1H), 3.59–3.61 (dd, J = 8 Hz, 2H), 3.26–3.32 (m, 4H), 2.88 (brs, 4H), 2.82 (s, 1H), 2.26 (brs, 6H); m/z: 340.35 (M = C17H26ClN3O2).3.5 Synthesis of (RS)-ranolazine (5)
To obtain the final drug molecule, (RS)-2-(4-(3-chloro-2-hydroxyphenyl)piperazin-1-yl)-N-(2,6-dimethylphenyl)acetamide 4 was further treated with methoxy phenol in the presence of K2CO3 in water which resulted the synthesis of (RS)-ranolazine 5 with 75% yield. Recovered product was characterized by GC-MS and NMR spectroscopy. 1H NMR (CDCl3, 400 MHz) δ: 6.98–6.96 (m, 3H), 6.86–6.77 (m, 4H), 4.06–4.02 (m, 1H), 3.89–3.86 (m, 1H),3.82–3.78 (m, 1H), 3.71 (s, 3H), 2.58 (brs, 8H), 2.55–2.63 (m, 4H) 2.25 (s, 6H); 13C NMR (CDCl3, 100 MHz) δ: 168.41, 149.4, 148.3, 134.97, 133.61, 128.32, 127.21, 121.97, 120.95, 114.78, 111.9, 72.3, 66.0, 61.66, 60.53, 55.8, 53.8, 53.58, 18.67; m/z: 428.38 (M = C24H33N3O4). The product was further characterized by chiral HPLC analysis using chiralcel OD-H column (hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2-propanol::9
2-propanol::9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), tR = 15 min, tS = 22.1 min.
1), tR = 15 min, tS = 22.1 min.
3.6 In silico study of screening of lipases for the kinetic resolution of (RS)-5
The docking study of (RS)-5 with different lipases was executed in the MVD programme using Docking wizard.38 In this process, initially eleven different lipases (2NW6, 3GUU, 1LPN, 1LPO, 1GZ7, 1CLE, 1UZA, 3LIP, 1PCN, 3TGL, 3W9B) were imported from PDB (Protein Data Bank) and complexed small molecules were removed. Prior to docking, the docking cavities were recognized using the ‘Cavity Prediction’ module which utilizes probe (size 1.20 Å) in a grid resolution of 0.80 Å, which expanded van-der-Waals molecular surface representation to specify docking site. MolDock scoring function and MolDock Optimizer genetic algorithm was employed to search and score dock poses. MolDock is an extended PLP based docking scoring function with newly added H-bonding and electrostatic energy terms to improve docking accuracy.39 The best dock poses were retrieved based on the lowest binding energy among multiple poses criteria and after graphical evaluations. The graphical illustrations of protein–ligand interactions were made using the Molegro Molecular Viewer (MMV) 2.5.0 programme (CLC Bio, Qiagen Inc.).40 Amino acid residues of lipases involved in the interaction with the ligand were determined and respective H-bond lengths were calculated. Also, this interaction with amino acid residues helped to explain the reaction mechanism. The differential interaction of (R)/(S)-5 with the lipases were shown through the Molegro Molecular Viewer and it reveals the basis for the enantioselectivity of the biocatalyst.3.7 Enantioselective transesterification of (RS)-5
Commercial lipases from different sources such as Candida antartica, C. rugosa 90860, C. rugosa 62316, C. rugosa L-1754, C. cylindracea, Aspergillus niger, porcine pancreas, AY-Amano 30 and the immobilized lipases like sol–gel-Ak from Pseudomonas cepacia, immobilized lipozyme from Mucor meihei, lipase acrylic resin from C. antartica CLEA, were used for kinetic resolution of (RS)-5. All the lipases were individually taken into different conical flasks (5 mL). Substrate (5) 5 mM in 0.8 mL toluene along with 0.2 mL vinyl acetate (2.5 mM), as acyl donor was added into each flask. The flasks were then capped and kept in an incubator shaker at 37 °C (200 rpm) for 48 h. Reactions were worked up after 48 h and the conversion and enantiomeric excess were analyzed by HPLC.The effect of different physico–chemical parameters such as organic solvents, reaction time, temperature, enzyme and substrate concentrations, and acyl donors was studied to optimize the reaction condition for achieving the best conversion and highest enantiomeric excess (see ESI S1–S3†).
3.8 Preparative-scale transesterification reaction of (RS)-5
The resolution of (RS)-5 was carried out in preparative scale under the optimized condition. The transesterification was performed with 5 mM substrate and C. antartica lipase A CLEA at 30 °C using vinyl acetate (2.5 mM) as acyl donor in 50 mL toluene. After 48 h, the reaction mixture was filtered and enzyme preparation was washed with toluene when the transformation was 48.5% with eeS of 80%. The solvent was evaporated under vacuum and resulting dried residue was subjected to flash chromatography (hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate::8
ethyl acetate::8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2).
2).
(S)-6: 1H NMR (CDCl3, 400 MHz) δ: 8.7–8.68 (s, 1H), 7.28 (brs, 3H), 7.0–6.91 (dd, 4H), 5.36 (s, 1H), 4.24–4.18 (m, 2H), 3.88–3.87 (d, J = 4 Hz, 3H), 3.24–3.20 (d, J = 16 Hz, 2H), 2.71–2.61 (m, 4H), 2.24 (brs, 6H), 2.09 (s, 2H), 1.59 (brs, 6H); m/z: 470.32 (M = C26H35N3O5). The product was further characterized by chiral HPLC using chiralcel OD-H column (hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2-propanol::9
2-propanol::9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), tS-6 = 13.12 min.
1), tS-6 = 13.12 min.
(R)-5: 1H NMR (CDCl3, 400 MHz) δ: 6.98–6.96 (m, 3H), 6.86–6.77 (m, 4H), 4.06–4.02 (m, 1H), 3.89–3.86 (m, 1H), 3.82–3.78 (m, 1H), 3.71 (s, 3H), 2.58 (brs, 8H), 2.55–2.63 (m, 4H), 2.25 (s, 6H); 13C NMR (CDCl3, 100 MHz) δ: 168.41, 149.4, 148.3, 134.97, 133.61, 128.32, 127.21, 121.97, 120.95, 114.78, 111.9, 72.3, 66.0, 61.66, 60.53, 55.8, 53.8, 53.58, 18.67; m/z: 428.38 (M = C24H33N3O4); [α]20D = +3.0 (c 4.0, CHCl3). The product was analyzed by HPLC using chiralcel OD-H column (hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2-propanol::9
2-propanol::9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), tR-5 = 15 min.
1), tR-5 = 15 min.
3.9 Deacylation of (S)-6 for the synthesis of (S)-5
A solution of K2CO3 (0.27 g, 2 mmol) in distilled water (1 mL) was added separately to (S)-6 (1 mmol) in methanol (5 mL) and the reaction mixture was kept under stirring for 2 h at room temperature (30 °C). After completion, the reaction mixture was extracted with EtOAc and water. The combined organic layer was dried over Na2SO4 and concentrated under vacuum to obtain the crude, which was purified by column chromatography (hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate::8
ethyl acetate::8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) on silica gel (100–200 mesh) to obtain (S)-5 (Scheme 3).
2) on silica gel (100–200 mesh) to obtain (S)-5 (Scheme 3).
(S)-5: 1H NMR (CDCl3, 400 MHz) δ: 6.98–6.96 (m, 3H), 6.86–6.77 (m, 4H), 4.06–4.02 (m, 1H), 3.89–3.86 (m, 1H), 3.82–3.78 (m, 1H), 3.71 (s, 3H), 2.58 (brs, 8H), 2.55–2.63 (m, 4H), 2.25 (s, 6H); 13C NMR (CDCl3, 100 MHz) δ: 168.41, 149.4, 148.3, 134.97, 133.61, 128.32, 127.21, 121.97, 120.95, 114.78, 111.9, 72.3, 66.0, 61.66, 60.53, 55.8, 53.8, 53.58, 18.67; m/z: 428.38 (M = C24H33N3O4); [α]20D = −3.1 (c 4.0, CHCl3). The product was further characterized by chiral HPLC using chiralcel OD-H column (hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2-propanol::9
2-propanol::9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), tS-5 = 22.1 min.
1), tS-5 = 22.1 min.
4. Conclusion
Computational catalysis has become a rationalized approach for in silico biocatalysts screening for the synthesis of enantiopure drugs and drug intermediates, as exemplified here using ranolazine as a model drug. The differential interaction of the two enantiomers with lipase, as determined by the docking score and H-bonding pattern, proved to be advantageous to select the suitable enzyme preparation for the kinetic resolution of (RS)-ranolazine. After the in silico and wet lab based screening, Candida antartica lipase CLEA was found to be the most efficient catalyst for the resolution of (RS)-ranolazine with higher conversion and enantiomeric excess. Under the optimized reaction conditions, 48.5% conversion with 80% enantiomeric excess was achieved which led towards the green synthesis of (S)-ranolazine. This combinatorial approach of in silico studies and biocatalysis to synthesize enantiopure ranolazine, which avoids the use of toxic metal catalysts, proved to be a game changer in biocatalysis.Acknowledgements
GS acknowledges Department of Biotechnology, Govt. of India for providing financial support to carry out the work. SG acknowledges Indian Council of Medical Research, Govt. of India for this work.References
- M. Höhne, S. Schätzle, H. Jochens, K. Robins and U. T. Bornscheuer, Nat. Chem. Biol., 2010, 6, 807–813 CrossRef PubMed.
- W. Li, B. Yanga, Y. Wang, D. Wei, C. Whiteley and X. Wanga, J. Mol. Catal. B: Enzym., 2009, 57, 299–303 CrossRef CAS.
- C. Orrenius, F. Hæffner, D. Rotticci, N. Öhrner, T. Norin and K. Hult, Biocatal. Biotransform., 1998, 16, 1–15 CrossRef CAS.
- F. Haeffner, T. Norin and K. Hult, Biophys. J., 1998, 74, 1251–1262 CrossRef CAS PubMed.
- R. A. Friesner, J. L. Banks, R. B. Murphy, T. A. Halgren, J. J. Klicic, D. T. Mainz, M. P. Repasky, E. H. Knoll, M. Shelley, J. K. Perry, D. E. Shaw, P. Francis and P. S. Shenkin, J. Med. Chem., 2004, 47, 1739–1749 CrossRef CAS PubMed.
- A. S. Bommarius and B. R. Riebel-Bommarius, Biocatalysis: Fundamentals and Applications, John Wiley & Sons, 2007, p. 634 Search PubMed.
- FDA's Policy statement for the development of new stereoisomeric drugs, Chirality, 1992, 4, 338–340 Search PubMed.
- A. Schmid, J. S. Dordick, B. Hauer, A. Kiener, M. Wubbolts and B. Witholt, Nature, 2001, 409, 258–268 CrossRef CAS PubMed.
- A. M. Thayer, ACS Chem. Eng. News, 2008, 86, 12–20 Search PubMed.
- M. Alcalde, M. Ferrer, F. J. Plou and A. Ballesteros, Trends Biotechnol., 2006, 24, 281–287 CrossRef CAS PubMed.
- S. Panke and M. Wubbolts, Curr. Opin. Chem. Biol., 2005, 9, 188–194 CrossRef CAS PubMed.
- R. N. Patel, Biomolecules, 2013, 3, 741–777 CrossRef PubMed.
- D. J. Pollard and J. M. Woodley, Trends Biotechnol., 2007, 25, 66–73 CrossRef CAS PubMed.
- D. R. Yazbeck, C. A. Martinez, S. Hu and J. Tao, Tetrahedron: Asymmetry, 2004, 15, 2757–2763 CrossRef CAS.
- E. P. Hudson, R. K. Eppler and D. S. Clark, Curr. Opin. Biotechnol., 2005, 16, 637–643 CrossRef CAS PubMed.
- B. P. Dwivedee, S. Ghosh, J. Bhaumik, L. Banoth and U. C. Banerjee, RSC Adv., 2015, 5, 15850–15860 RSC.
- S. Ghosh, J. Bhaumik, L. Banoth, S. Banesh and U. C. Banerjee, Chirality, 2016, 28, 313–318 CrossRef CAS PubMed.
- N. Bouzemi, H. Debbeche, L. Aribi-Zouioueche and J. C. Fiaud, Tetrahedron Lett., 2004, 45, 627–630 CrossRef CAS.
- J. Köhler and B. Wünsch, Theor. Biol. Med. Modell., 2007, 4, 34 CrossRef PubMed.
- FDA labelling information, (a) http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/2006/ucm108587.htm; (b) http://www.accessdata.fda.gov/drugsatfda_docs/label/2008/021526s004lbl.pdf.
- S. Aalla, G. Gilla, R. R. Anumula, S. Kurella, P. R. Padi and P. R. Vummenthala, Org. Process Res. Dev., 2012, 16, 748–754 CrossRef CAS.
- Heart disease and stroke statistics—2015 update: a report from the American Heart Association, Circulation, DOI:10.1161/CIR.0000000000000152.
- T. Reffelmann and R. A. Kloner, Expert Rev. Cardiovasc. Ther., 2010, 8, 319–329 CrossRef CAS PubMed.
- S. Sossalla and L. S. Maier, Pharmacol. Ther., 2012, 133, 311–323 CrossRef CAS PubMed.
- J. G. McCormack, W. C. Stanley and A. A. Wolff, Gen. Pharmacol. Vasc. Syst., 1998, 30, 639–645 CrossRef CAS.
- A. Dhalla, K. Leung, J. Shryock and D. Zeng, PCT/US2008/060090, 2008.
- S. Li, Chin. J. Med. Chem., 2003, 13, 283–285 CAS.
- W. Lu, et al., Zhongguo Yiyao Gongye Zazhi, 2004, 35, 641–642 CAS.
- S. Aalla, G. Gilla, R. R. Anumula, S. Kurella, P. R. Padi and P. R. Vummenthala, Org. Process Res. Dev., 2012, 16, 748–754 CrossRef CAS.
- M. A. Riise, R. Karstad and T. Anthonsen, Biocatal. Biotransform., 2005, 23, 45–51 CrossRef.
- I. C. Gutierrez and R. X. Garcia, US 2009/0318697A1, 2009.
- D. N. Kommi, D. Kumar and A. K. Chakraborti, Green Chem., 2013, 15, 756–767 RSC.
- S. P. Pathare and K. G. Akamanchi, Tetrahedron Lett., 2013, 54, 6455–6459 CrossRef CAS.
- N. Gurung, S. Ray, S. Bose and V. Rai, BioMed Res. Int., 2013, 1–18 CrossRef PubMed.
- S. G. Estacio, Enzymology: Bringing Together Experiments and Computing, 2012, vol. 87, p. 249 Search PubMed.
- A. Jongejan, C. de Graaf, N. P. Vermeulen, R. Leurs and I. J. de Esch, Methods Mol. Biol., 2005, 310, 63–91 CAS.
- R. Huber and W. S. Bennett, Biopolymers, 1983, 22, 261–279 CrossRef CAS PubMed.
- Molegro Virtual Docker (MVD), CLC Drug Discovery Workbench 1.0, CLC Bio, Qiagen Inc., 2014, software available at http://www.clcbio.com/products/clc-drug-discoveryworkbench Search PubMed.
- R. Thomsen and M. H. Christensen, MolDock, J. Med. Chem., 2006, 49, 3315–3321 CrossRef CAS PubMed.
- Molegro Molecular Viewer (MMV) 2.5.0, CLC Bio, Qiagen Inc., 2012, software available at http://www.clcbio.com/products/clc-drug-discovery-workbench Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra06879k |
| This journal is © The Royal Society of Chemistry 2016 |




