Determination of the secondary structure of peptides in the presence of Gram positive bacterium S. epidermidis cells†
C. Avitabilea,
L. D. D'Andreaa,
M. Savianob and
A. Romanelli*ac
aIstituto di Biostrutture e Bioimmagini, CNR, via Mezzocannone 16, 80134 Napoli, Italy
bIstituto di Cristallografia, CNR, via Amendola 122, 70126 Bari, Italy
cDipartimento di Farmacia, Università di Napoli “Federico II”, via Mezzocannone 16, 80134 Napoli, Italy. E-mail: alessandra.romanelli@unina.it
First published on 20th May 2016
Abstract
A protocol to run CD experiments in the presence of Gram positive bacterial cells was set up. The secondary structure of the antimicrobial peptides temporin L and TB_KKG6A with S. epidermidis cells was determined. CD experiments were also reported for temporin L in the presence of Gram negative bacterium E. coli cells.
The antimicrobial activity of peptides seems to be mediated by the interactions of peptides with the bacterial cell envelope. Recent studies have been performed to clarify details at the molecular level on the interactions of antimicrobial peptides with the Gram negative bacterium E. coli.1 Initial studies carried out on cyclic R-, W-rich peptides tested on E. coli LPS-mutant phenotypes showed that the interaction with the lipopolysaccharide (LPS), which is the main component of the Gram negative bacteria outer membrane, is crucial for the selectivity of antimicrobial peptides (AMPs) on E. coli cells.2 It has been demonstrated by AFM measurements on E. coli cells that the antimicrobial peptides BP100 and pepR cause the collapse of bacterial cells at a concentration equal to the MIC. In these conditions the peptides cover the bacterial surface and neutralize its charge.3 Time-lapse fluorescence lifetime imaging has been employed to monitor E. coli membrane disruption by a melittin analogue: transient pore formation was observed in E. coli cells, in contrast to stable pore formation which occurred in artificial membranes.4 Time-resolved flow cytometry has been employed to monitor the pepR induced permeabilization on three E. coli strains: these studies demonstrate that the LPS composition affects the membrane permeabilization events and the bacteria killing mediated by antimicrobial peptides.5 In some cases it is known that the interactions of antimicrobial peptides with Gram negative bacterial cells result in the folding of peptides. Recent experiments carried out in our lab have demonstrated that, upon interaction with the E. coli cells, peptides such as cecropin A, magainin 2 and the analogue of temporin B TB_KKG6A assume an alpha helical conformation.6,7 In these cases the LPS seems to trigger the folding of peptides. However, the three-dimensional structure of the peptide TB_KKG6A determined in the presence of E. coli LPS and cells as well suggests that interactions of the peptide with components other than the LPS located on the bacterial outer membrane affect the peptide structure. Interactions of AMPs with Gram positive cells components are very little investigated. The composition of Gram positive bacteria envelope is very complex.8 Gram positive bacteria lack a protective outer membrane, which instead protects Gram negative bacteria. A layer of peptidoglycan 30–100 nm thick surrounds Gram positive microorganisms and is also the anchor for surface proteins, which may be covalently attached or non-covalently associated to the peptidoglycan. Wall teichoic acids (WTA) are connected to the peptidoglycan through a phosphodiester bond, while lipoteichoic acids (LTA) are attached to glycolipids embedded in the membrane. The composition of the cell envelope, as well as the localization of the envelope components are functional to the selective intake of nutrients, expulsion of waste and to the survival of bacteria in hostile environments. Interaction of antimicrobial peptides active against Gram positive bacteria with lipid II has been hypothesized in a number of cases. Lipid II is a precursor of the cell wall; it contains a subunit of the peptidoglycan bound to the membrane poly-isoprenoid by a pyrophosphate linker.9 It is reported that defensins such as the fungal plectasin, the human β-defensin 3 and α-defensin 1 but also the lantibiotic nisin and vancomycin bind lipid II and inhibit cell wall biosynthesis.10 Peptides may either form pores in the membrane, causing its lysis or may remove lipid II from the cell division site. In both cases cell wall synthesis is blocked. The binding sites of peptides on lipid II are the anionic diphosphates for lantibiotics and defensin and the peptide chains for vancomycin. Changes in the three-dimensional structure of peptides occur upon binding to lipid II: for example the peptide mersacidin exposes its charged residues in the presence of lipid II.11 Few studies are focused on the structure of AMPs in the presence of lipid II and, as far as we know, there are no structural studies carried out in the presence of Gram positive bacterial cells. These studies will allow the gaining of new details on the mechanism of action of antimicrobial peptides, yielding information on the relationship between the structure and the activity of peptides. In this work we describe a protocol to detect the secondary structure of antimicrobial peptides in the presence of Gram positive bacterial cells. We report circular dichroism studies on two peptides belonging to the temporin family, temporin L (TL) and TB_KKG6A, in the presence of S. epidermidis cells. Temporin L is a natural peptide, TB_KKG6A is an analogue of temporin B recently developed in our lab; both peptides are active against Gram positive and Gram negative bacteria.12–14 For our experiments we choose the S. epidermidis strain ATCC 12228, as this is a non-pathogenic and non-biofilm forming strain and can easily be handled in the lab.15 In order to compare the conformation of the peptides on Gram positive and Gram negative bacteria, we determined the secondary structure of the peptide TL in the presence of the Gram negative bacteria E. coli. Initial efforts were devoted at setting up the conditions for our experiments; in particular the choice of a buffer compatible with either cell survival and CD experiments is crucial. S. epidermidis cells, unlike E. coli cells, shrink if incubated in phosphate buffer 10 mM, at pH 7. When incubated in a Na2HPO4 10 mM, KH2PO4 2 mM, NaCl 20 mM, KCl 2.7 mM buffer at pH 7, S. epidermidis cells survive for at least two hours. The optimal cell and peptide concentrations for running CD experiments were chosen based on the results that we obtained on the peptide + cell mixtures, exploring cell concentrations ranging from 0.08 to 0.1 OD600 and peptide concentrations ranging from 5 to 30 μM. CD spectra were recorded in all cases using cells at 0.1 OD600 nm; in these conditions the CD spectra of cells are quite flat, showing a negative signal whose intensity is always in the range −1 to −3 CD mdeg (Fig. 1).
 | ||
| Fig. 1 CD spectra of S. epidermidis cells 0.1 OD600 after 20 minutes (green), 60 minutes (green), 80 minutes (magenta) in buffer. These spectra are representative of one set of experiments. | ||
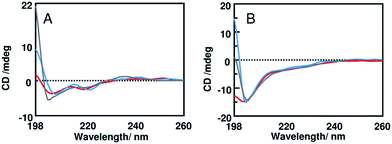
Measurements were carried out every 20 minutes for the first two hours and every two hours up to six hours. In all cases we recorded the spectra of cells and the spectra of mixtures peptide + cells. Subtraction of the CD spectrum of cells to the CD spectrum of the cells + peptide mixture affords the CD spectrum of the peptide. We assume that the spectrum of cell does not change drastically due to the presence of the peptides. In all cases explored, the intensity of the CD spectrum of cells is very low in the range 200–260 nm and in fact CD spectra of the mixtures peptides + cells look very similar to the subtracted spectra (Fig. S1†). Both peptides have not structural preferences in phosphate buffer, as elsewhere reported.7,16 CD spectra recorded for TL incubated with S. epidermidis cells indicate that this peptide folds into an alpha helix: spectra recorded in the time range 20–60 minutes show two negative bands around 220 and 206 nm (Fig. 2A). After 80 minutes of incubation, the intensity of the band at 220 nm decreases and the other band shifts to 203 nm; then CD spectra keep changing with the time (Fig. S2†). The secondary structure of TB_KKG6A in the presence of S. epidermidis cells is different from that observed for TL. Upon incubation with cells, the CD spectrum of TB_KKG6A shows a very weak negative band around 225 nm, a minimum at 203 nm (Fig. 2B). The appearance of the spectrum doesn't change significantly within 4 hours. After 6 hours the CD spectrum does not show any cross at 200 nm, a defined band at 228 nm appears (Fig. S2†). Association of this band to a conformation of the peptide is at the moment very hard. CD bands at this wavelength have in fact been reported for peptides containing aromatic residues, which are absent in TB_KKG6A.2,17 Unlike TL, TB_KKG6A seems to be mostly unfolded in the presence of S. epidermidis cells.
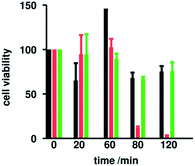
When we compare the vitality of S. epidermidis cells, measured after counting the CFU mL−1, in the presence of the peptides TB_KKG6A and TL, in the same conditions as those employed to run CD experiments, we see that up to sixty minutes bacterial cells are still vital (Fig. 3). These data confirm that in this time frame we are looking at the conformation of peptides in the presence of live cells. After 80 minutes, all cells start to die. Killing kinetics look different and it is likely that the two peptides act by different mechanisms. The change of the CD spectra of TL after 80 min of incubation with S. epidermidis cells is in line with the viability experiments and might be due to the fragmentation of the bacterial envelope. If the surface with which the peptide interacts is not intact and homogeneous anymore, it is likely to see changes in the secondary structure of the peptide.
The fact that the spectra of TB_KKG6A appear the same for about 4 hours, might be an indication of the fact the peptide is always in the same environment. This, in turn, may be due to the fact that fragmentation of the cell envelope does not occur. We also recorded spectra at lower peptide concentrations (5 μM for TB_KKG6A and 10 μM for TL) and 0.1 OD600, to be sure that we're capturing the conformations of the peptides when they are interacting with live bacteria. As expected, these spectra (reported in Fig. S3 and S4†) are less intense as compared to those reported in Fig. 2 but have the same shape. Assuming that interaction of peptides with bacterial cell envelope components is crucial for activity, we can conclude that TL and TB_KKG6A need to adopt different conformations in order to maximize the interaction. It is also likely that the different structure of the two peptides on S. epidermidis cells is due to the fact that the peptides interact with different partner on the cell envelope.
On the other side, we observe that TL and TB_KKG6A assume a defined alpha helical conformation in the presence of E. coli cells. The secondary and three-dimensional and structure of TB_KKG6A has been previously reported.7 The analysis of the secondary structure of TL in the presence of E. coli cells is here described. The peptide TL folds within 60 minutes, CD spectra show two bands at 223 and 209 nm, which clearly suggest that TL folds into an α alpha helix (Fig. 4). The NMR structure reported for TL in the presence of E. coli LPS shows that this peptide forms an antiparallel dimer, composed of two helices.18 Dimerization affords a perfectly amphipathic structure which exposes a positively charged surface, likely devoted to the interaction with the negatively charged phosphates of the lipopolysaccharide lipid A and a hydrophobic surface which has been hypothesized to establish interactions with the acyl chain of LPS. It is likely that TL assumes a similar conformation also with E. coli cells. The appearance of the spectra of TL + E. coli cells does not change after two hours of incubation of the peptide with E. coli cells (Fig. 4). When the same experiments were carried out on TB_KKG6A, a change in the position and intensity of the CD bands was observed after 80 minutes.7 This difference might be ascribed to a quicker disruption of Gram negative bacterial membrane by TB_KKG6A as compared to TL, which might determine aggregation or precipitation of the peptides bound to membrane fragments, and consequently a decrease in the concentration of helical folded peptides in solution. Results obtained for TL and E. coli cells appear reasonable in light of data reported by Mangoni et al. showing that temporin L does not lyse E. coli cells, nor produces holes in the membranes but rather induces formation of ghost-like bacteria.19 The cell wall of the bacteria remains intact and therefore the peptide is always in the same environment.
Conclusions
We have demonstrated that interaction of the antimicrobial peptides TL and TB_KKG6A with the cell envelope of S. epidermidis results in different secondary structures for the two peptides. TL assumes an alpha helical conformation in the presence of Gram positive and Gram negative bacterial cells, while TB_KKG6A assumes a defined conformation only in the presence of E. coli cells. These observations strongly support the idea that the two peptides interact in a different way with bacteria. It has to be cleared if the different structures observed for the two peptides as analyzed with S. epidermidis cells and also the different evolution of the CD spectra of peptides in the presence of E. coli cells within the time imply different mechanism of action.Notes and references
- J. M. Freire, D. Gaspar, A. S. Veiga and M. A. Castanho, J. Pept. Sci., 2015, 21, 178–185 CrossRef CAS PubMed.
- C. Junkes, R. D. Harvey, K. D. Bruce, R. Dolling, M. Bagheri and M. Dathe, Eur. Biophys. J., 2011, 40, 515–528 CrossRef CAS PubMed.
- C. S. Alves, M. N. Melo, H. G. Franquelim, R. Ferre, M. Planas, L. Feliu, E. Bardaji, W. Kowalczyk, D. Andreu, N. C. Santos, M. X. Fernandes and M. A. Castanho, J. Biol. Chem., 2010, 285, 27536–27544 CrossRef CAS PubMed.
- M. L. Gee, M. Burton, A. Grevis-James, M. A. Hossain, S. McArthur, E. A. Palombo, J. D. Wade and A. H. Clayton, Sci. Rep., 2013, 3, 1557 Search PubMed.
- J. M. Freire, D. Gaspar, B. G. de la Torre, A. S. Veiga, D. Andreu and M. A. Castanho, Biochim. Biophys. Acta, 2015, 1848, 554–560 CrossRef CAS PubMed.
- C. Avitabile, L. D. D'Andrea and A. Romanelli, Sci. Rep., 2014, 4, 4293 Search PubMed.
- G. Malgieri, C. Avitabile, M. Palmieri, L. D. D'Andrea, C. Isernia, A. Romanelli and R. Fattorusso, ACS Chem. Biol., 2015, 10, 965–969 CrossRef CAS PubMed.
- T. J. Silhavy, D. Kahne and S. Walker, Cold Spring Harbor Perspect. Biol., 2010, 2, a000414 Search PubMed.
- E. Breukink and B. de Kruijff, Nat. Rev. Drug Discovery, 2006, 5, 321–332 CrossRef CAS PubMed.
- D. Munch and H. G. Sahl, Biochim. Biophys. Acta, 2015, 1848, 3062–3071 CrossRef PubMed.
- S. T. D. Hsu, E. Breukink, G. Bierbaum, H. G. Sahl, B. de Kruijff, R. Kaptein, N. A. J. van Nuland and A. M. J. J. Bonvin, J. Biol. Chem., 2003, 278, 13110–13117 CrossRef CAS PubMed.
- A. C. Rinaldi, M. L. Mangoni, A. Rufo, C. Luzi, D. Barra, H. Zhao, P. K. Kinnunen, A. Bozzi, A. Di Giulio and M. Simmaco, Biochem. J., 2002, 368, 91–100 CrossRef CAS PubMed.
- C. Avitabile, F. Netti, G. Orefice, M. Palmieri, N. Nocerino, G. Malgieri, L. D. D'Andrea, R. Capparelli, R. Fattorusso and A. Romanelli, Biochim. Biophys. Acta, 2013, 1830, 3767–3775 CrossRef CAS PubMed.
- V. Bezzerri, C. Avitabile, M. C. Dechecchi, I. Lampronti, M. Borgatti, G. Montagner, G. Cabrini, R. Gambari and A. Romanelli, J. Pept. Sci., 2014, 20, 822–830 CrossRef CAS PubMed.
- Y. Q. Zhang, S. X. Ren, H. L. Li, Y. X. Wang, G. Fu, J. Yang, Z. Q. Qin, Y. G. Miao, W. Y. Wang, R. S. Chen, Y. Shen, Z. Chen, Z. H. Yuan, G. P. Zhao, D. Qu, A. Danchin and Y. M. Wen, Mol. Microbiol., 2003, 49, 1577–1593 CrossRef CAS PubMed.
- A. Carotenuto, S. Malfi, M. R. Saviello, P. Campiglia, I. Gomez-Monterrey, M. L. Mangoni, L. M. Gaddi, E. Novellino and P. Grieco, J. Med. Chem., 2008, 51, 2354–2362 CrossRef CAS PubMed.
- P. L. Clark, Z. P. Liu, J. Zhang and L. M. Gierasch, Protein Sci., 1996, 5, 1108–1117 CrossRef CAS PubMed.
- A. Bhunia, R. Saravanan, H. Mohanram, M. L. Mangoni and S. Bhattacharjya, J. Biol. Chem., 2011, 286, 24394–24406 CrossRef CAS PubMed.
- M. L. Mangoni, N. Papo, D. Barra, M. Simmaco, A. Bozzi, A. Di Giulio and A. C. Rinaldi, Biochem. J., 2004, 380, 859–865 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures, additional CD data. See DOI: 10.1039/c6ra06877d |
| This journal is © The Royal Society of Chemistry 2016 |