Production of GDP-l-fucose from exogenous fucose through the salvage pathway in Mortierella alpina†
Abstract
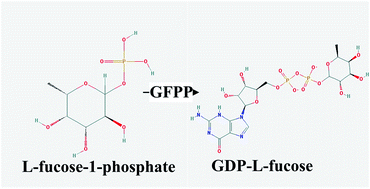
GDP-L-fucose is an essential donor for the biosynthesis of fucosyloligosaccharides, participating in a variety of biological and pathological processes. Mortierella alpina can accumulate lipids in quantities up to 50% of its dry weight and has been proved to possess a GDP-L-fucose de novo synthesis pathway. Analysis of the M. alpina genome suggests that there is a putative GDP-L-fucose pyrophosphorylase (GFPP) gene playing a role in the salvage pathway of GDP-L-fucose, which has never been found in fungi before. To explore the molecular mechanisms of salvage reactions for free fucose in fungi, GFPP was expressed heterologously in Escherichia coli and the recombinant enzyme was purified to homogeneity. The enzymatic activity was investigated by liquid chromatography and mass spectrometry. Characterization of the GFPP in M. alpina indicated this fungus can convert fucose to GDP-L-fucose through the salvage pathway. The addition of fucose at the initial stage of cell multiplication exerted no impact on the GDP-L-fucose content in cells, whereas the addition of fucose (10 nM) after nitrogen exhaustion led to an increase of GDP-L-fucose production by 50%. Furthermore, medium supplementation with a combination of fucose and Mg2+ (10 nM) led to a 2.7-fold increase in the yield of GDP-L-fucose in M. alpina (0.57 mg per g cell). Additionally, the transcript level of GFPP is upregulated by the addition of fucose and Mg2+, which highlights the functional significance of GFPP in GDP-L-fucose biosynthesis. To our knowledge, this study is the first to report a comprehensive characterization of GFPP in a fungus.

 Please wait while we load your content...
Please wait while we load your content...