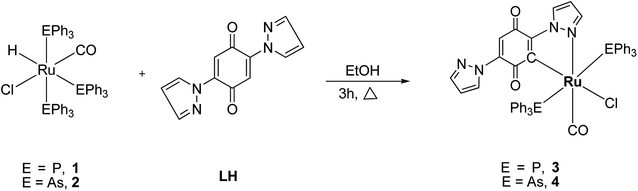
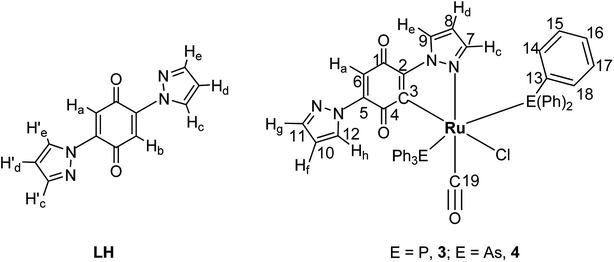
Impact of chelation on anticancer activities of organometallic ruthenium(II) complexes containing 2,5-di(1H-pyrazol-1-yl)-1,4-benzoquinone: synthesis, structure, DNA/protein binding, antioxidant activity and cytotoxicity†
Thangavel Sathiya Kamatchiad,
Palaniappan Kalaivanib,
Frank R. Fronczekc,
Karuppannan Natarajan*a and
Rathinasabapathi Prabhakaran*a
aDepartment of Chemistry, Bharathiar University, Coimbatore 641046, India. E-mail: k_natraj6@yahoo.com; rpnchemist@gmail.com; Fax: +91 422 2422387; Tel: +91 422 2428319
bDepartment of Chemistry, Nirmala College for Women, Bharathiar University, Coimbatore 641018, India
cDepartment of Chemistry, Louisiana State University, Baton Rouge, LA 70803, USA
dDepartment of Chemistry, Sakthi College of Arts and Science for Women, Oddanchatram, Dindigul 624 624, India
First published on 3rd May 2016
Abstract
To gain some insight into the influence of chelation on anticancer activities, two new bivalent organometallic ruthenium complexes, [Ru(L)Cl(CO)(PPh3)2] (3) and [Ru(L)Cl(CO)(AsPh3)2] (4), (HL = 2,5-di(1H-pyrazol-1-yl)-1,4-benzoquinone) were synthesized, structurally characterized comprehensively by elemental analysis, IR, electronic, 1H/13C NMR spectral studies and their biological activities (DNA/protein interactions, antioxidant and cytotoxic activity studies) have been investigated and compared with that of appropriate precursor complexes [RuHCl(CO)(PPh3)3] (1), [RuHCl(CO)(AsPh3)3] (2) and the ligand LH. The crystal structures of both the new complexes 3 and 4 were determined by single crystal X-ray diffraction studies, which unveil that the central ruthenium ion adopted distorted octahedral geometry with L as a monobasic bidentate donor and the chelator was observed to undergo C–H activation at one of the ortho positions leading to the formation of a five-membered metallacycle. DNA/protein interactions of the complexes have been examined by photophysical studies, which demonstrated a non-intercalative binding mode of the complexes with DNA and the complexes strongly quench the fluorescence intensity of bovine serum albumin (BSA) through the mechanism of static quenching. The new complexes demonstrated better binding affinity with DNA/protein. The compounds were evaluated for their free radical scavenging ability involving DPPH radicals, hydroxyl radicals, nitric oxide radicals, superoxide anion radicals and hydrogen peroxide. The results showed that the new complexes 3 and 4 possess significant radical scavenging ability and outperform the standard antioxidants vitamin C and BHT. The results of in vitro cytotoxicity of the complexes tested against B16F10, HeLa, A549, MCF7 and SKOV3 cells by SRB assay were compared with the standard drug cisplatin and that the new complexes 3 and 4 showed better activity in inhibiting the growth of the cancer cells. The influence of chelation was seen in all the biological activities studied and the new ruthenium chelates 3 and 4 were found to be superior to the precursor complexes 1, 2 and the ligand LH in DNA/protein binding properties, radical scavenging ability and cytotoxicity.
Introduction
Metal ions play a vital role in a vast number of biological processes1,2 and it is known that chelation of metal ions with organic ligands acts synergistically to increase their biological activities.3,4 Further, many organic compounds used in medicine do not have a purely organic mode of action and require traces of metal ions directly or indirectly for activation or biotransformation. Since antiquity, human beings have suffered from a variety of diseases out of which cancer is the second most common cause of death after cardiovascular diseases. In the search for more selective anti-tumour agents, there is considerable interest in finding metal complexes as bio-reductive drugs. Bio-reductive drugs are designed to take advantage of some of the unique features of solid tumours, in particular reduced oxygen tension and the over-expression of certain reductase enzymes,5 which are capable of reducing several quinone-containing anticancer agents to DNA-damaging species.6–8 The biological activity of the quinones is attributed to their facile in vivo and in vitro reduction.9 It is well known that any change in the substituent attached to the quinone ring alters its capability to accept electrons by tuning its redox chemistry and consequently modifies its biological activity.10–12 In addition to quinones, different pyrazole derivatives have also been tested for their antiproliferative activities in vitro and antitumor activity in vivo, often resulting in promising lead compounds.13–17 Moreover, a variety of 1,4-benzoquinones and their nitrogen analogues have been reported for their anti-tumour activities.18–20 The sensible to design the bioactive ligand was to combine in a single molecule, the known pharmacophore benzoquinone ring with anchor pyrazole ring. On moving to the bio-reductive metal fragment to chelate pyrazole bound benzoquinone ligand, ruthenium scaffolds belong to a very promising class of anti-tumor agents, which have an important role of the oxidation state that allows them to exist in the biological fluids in almost all common oxidation states from II to IV.21 A number of ruthenium complexes containing heterocyclic rings have previously been shown to display promising anticancer activities, and two of them, NAMI-A and KP109, have been scheduled for clinical trials.22–24With this back ground in mind, we attempted to synthesize new ruthenium chelates by reacting [RuHCl(CO)(PPh3)3]/[RuHCl(CO)(AsPh3)3] and 2,5-di(1H-pyrazol-1-yl)-1,4-benzoquinone and obtained new organometallic ruthenium(II) complexes. Here in, we report the synthesis, structure, DNA/protein binding, free radical scavenging and cytotoxicity of new novel organometallic ruthenium(II) complexes. Though 2,5-di(1H-pyrazol-1-yl)-1,4-benzoquinone is known to organic chemists as intermediates, it has not been used as a ligand for the synthesis of any metal complexes.
Experimental section
Materials and instrumentation
All the chemicals were reagent grade and were used as received from commercial suppliers unless otherwise stated. All the solvents were degassed and distilled according to the standard procedures.25 Commercially available RuCl3·3H2O (Himedia) was used to prepare the precursor complexes. The starting complexes [RuHCl(CO)(PPh3)3],26 [RuHCl(CO)(AsPh3)3]27 were prepared as reported earlier. The ligand 2,5-di(1H-pyrazol-1-yl)-1,4-benzoquinone (LH) was prepared according to the literature procedure28 with necessary modifications. Melting points were determined with Lab India instrument. Elemental analyses of carbon, hydrogen and nitrogen were performed on Vario EL III Elementar elemental analyzer. Electronic absorption spectra of the complexes were recorded using JASCO 600 spectrophotometer and emission measurements were carried out by using a JASCO FP-6600 spectrofluorometer. Nicolet Avatar Model FT-IR spectrophotometer was used to record the IR spectra (4000–400 cm−1) of the free ligand and the complexes as KBr pellets. 1H NMR and 13C NMR spectra were recorded on Bruker AV 500 (500 MHz (1H) and 125 MHz (13C)) spectrometer using tetramethylsilane (TMS) as an internal reference. The chemical shifts are expressed in parts per million (ppm). Protein free herring sperm DNA obtained from SRL chemicals was stored at 0–4 °C. Bovine serum albumin (BSA) and ethidium bromide (EB) were obtained from Sigma-Aldrich. Antioxidant activity measurements were done using UV spectrophotometer (UV-1800, Shimadzu).X-ray crystallography
X-ray diffraction measurements were performed on a Nonius Kappa CCD diffractometer equipped with graphite monochromated Mo Kα radiation and an Oxford Cryostream cryostat. The structure of the complexes was solved by direct methods and refinements were carried out by full matrix least-squares techniques. The hydrogen atoms were generally visible in difference maps and were placed in idealized positions and treated as riding in the refinement. The following computer programs were used: structure solution SIR-97,29 refinement SHELXL-97,30 molecular diagrams and ORTEP-3 (ref. 31) for Windows.DNA interaction studies
All the experiments involving the binding of the compounds with HS-DNA were carried out in deionised water with tris(hydroxymethyl)-aminomethane (Tris, 5 mM) and sodium chloride (50 mM) and adjusted to pH 7.2 with hydrochloric acid at room temperature. A solution of herring sperm DNA (0.1 g) in Tris–HCl buffer (10 mL) gave a ratio of UV absorbance at 260 and 280 nm of ca. 1.7–1.8: 1 indicating that the DNA was sufficiently free of protein. The DNA concentration per nucleotide was determined by absorption spectroscopy using the molar absorption coefficient (6600 M−1 cm−1) at 260 nm. The compounds were dissolved in a mixed solvent of 5% DMSO and 95% Tris–HCl buffer. Absorption spectra were recorded after equilibrium at 20 °C for 10 min. Absorption titration experiments were performed with a fixed concentration of the compounds (10 μM) while gradually increasing the concentration of DNA (0–50 μM). While measuring the absorption spectra, an equal amount of DNA was added to both the test solution and the reference solution to eliminate the absorbance of DNA itself.In order to know the exact mode of attachment of compounds with HS-DNA, fluorescence quenching experiments of ethidium bromide (referred to as EB)-DNA complex were also carried out by increasing concentration of ruthenium(II) complexes (0–50 μM) to the samples containing 10 μM EB, 10 μM DNA and Tris buffer. Before the measurements, the system was shaken and incubated at room temperature for ∼5 min. The emission was recorded at 530–750 nm. The fluorescence spectra were obtained with an excitation wavelength of 522 nm and an emission wavelength of 620 nm.
Protein binding studies
Binding of the compounds with bovine serum albumin (BSA) was studied from the fluorescence spectra recorded with an excitation wavelength of at 280 nm and the corresponding emission at 345 nm assignable to that of BSA. The excitation and emission slit widths and scan rates were maintained constant for all of the experiments. A stock solution of BSA was prepared in 50 mM phosphate buffer (pH = 7.2) and stored in the dark at 4 °C for further use. A concentrated stock solution of the compounds was prepared as mentioned for the DNA binding experiments, except that the phosphate buffer was used instead of a Tris–HCl buffer for all of the experiments. Titrations were manually done by using a micropipette for the addition of the compounds. For synchronous fluorescence spectra also, the same concentrations of BSA and the compounds were used, and the spectra were measured at two different Δλ values (difference between the excitation and emission wavelengths of BSA), such as 15 and 60 nm.Antioxidant assays
The ability of the ligand and the new organometallic ruthenium complexes to act as hydrogen donors or free radical scavengers was tested by conducting a series of in vitro antioxidant assays involving DPPH radical, hydroxyl radical, nitric oxide radical, hydrogen peroxide, superoxide anion radical and metal chelating assay. The results obtained were compared with standard antioxidants including natural antioxidant vitamin C and synthetic antioxidant BHT (Butylated Hydroxy Toluene).DPPH˙ scavenging assay
The DPPH radical scavenging activity of the compounds was measured according to the method of Blois.32 DPPH radical is a stable free radical and due to the presence of an odd electron, it shows a strong absorption band at 517 nm in visible spectrum. If this electron becomes paired off in the presence of a free radical scavenger, this absorption vanishes resulting in decolorization stoichiometrically with respect to the number of electrons taken up. Various concentrations of the experimental compounds were taken and the volumes were adjusted to 100 μL with methanol. About 5 mL of 0.1 mM methanolic solution of DPPH was added to the portion of samples and standards (BHT and vitamin C) and shaken vigorously. Negative control was prepared by adding 100 μL of methanol in 5 mL of 0.1 mM methanolic solution DPPH. The tubes were allowed to stand for 20 minutes at 27 °C. The absorbance of the sample was measured at 517 nm against the blank (methanol).OH˙ scavenging assay
The hydroxyl radical scavenging activity of the compounds has been investigated by using the Nash method.33 In vitro hydroxyl radicals were generated by Fe3+/ascorbic acid system. The detection of hydroxyl radicals was carried out by measuring the amount of formaldehyde formed from the oxidation reaction with DMSO. The formaldehyde produced was detected spectrophotometrically at 412 nm. In a typical experiment, a mixture of 1.0 mL of iron–EDTA solution (0.13% ferrous ammonium sulfate and 0.26% EDTA), 0.5 mL of EDTA solution (0.018%), and 1.0 mL of DMSO (0.85% DMSO (v/v) in 0.1 M phosphate buffer, pH 7.4) were sequentially added in the test tubes which contains fixed concentration of the test compounds. The reaction was initiated by adding 0.5 mL of ascorbic acid (0.22%) and was incubated at 80–90 °C for 15 min in a water bath. After incubation, the reaction was terminated by the addition of 1.0 mL of ice-cold trichloroacetic acid (17.5% w/v). Subsequently, 3.0 mL of Nash reagent was added to each tube and left at room temperature for 15 min. The intensity of the colour formed was measured spectrophotometrically at 412 nm against the reagent blank.NO˙ scavenging assay
Assay of nitric oxide (NO˙) scavenging activity is based on the method,34 where sodium nitroprusside in aqueous solution at physiological pH spontaneously generates nitric oxide, which interacts with oxygen to produce nitrite ions. This can be estimated using Griess reagent. Scavengers of nitric oxide compete with oxygen leading to reduced production of nitrite ions. For the experiment, sodium nitroprusside (10 mM) in phosphate buffered saline was mixed with a fixed concentration of the compounds, standards and incubated at room temperature for 150 min. After the incubation period, 0.5 mL of Griess reagent containing 1% sulfanilamide, 2% H3PO4 and 0.1% N-(1-naphthyl)ethylenediaminedihydrochloride was added. The absorbance of the chromophore formed was measured at 546 nm.H2O2 scavenging assay
The ability of the compounds to scavenge hydrogen peroxide was determined using the method of Ruch et al.35 In a typical experiment, a solution of hydrogen peroxide (2.0 mM) was prepared in phosphate buffer (0.2 M, pH-7.4) and its concentration was determined spectrophotometrically from absorption at 230 nm with molar extinction coefficient 81 M−1 cm−1. The compounds (100 μg mL−1), BHT and vitamin C (100 μg mL−1) were added to 3.4 mL of phosphate buffer prepared above together with hydrogen peroxide solution (0.6 mL). An identical reaction mixture without the sample was taken as negative control. Absorbance of hydrogen peroxide at 230 nm was determined after 10 min against the blank (phosphate buffer).O2−˙ scavenging assay
The superoxide anion radical (O2−˙) scavenging assay is based on the capacity of the compounds to inhibit formazan formation by scavenging the superoxide radicals generated in riboflavin–light–NBT system.36 In a typical experiment, a 3 mL reaction mixture contained 50 mM sodium phosphate buffer (pH 7.6), 20 μg riboflavin, 12 mM EDTA, 0.1 mg NBT and 1 mL complex solution (20–100 μg mL−1). Reaction was started by illuminating the reaction mixture with different concentrations of complex for 90 s and immediately, the absorbance was measured at 590 nm. The entire reaction assembly was enclosed in a box lined with aluminium foil. Identical tubes with reaction mixture kept in dark served as blanks.For the above five assays, all the tests were run in triplicate and the percentage activity was calculated using the following equation
| Scavenging activity (%) = [(A0 − A1)/A0] × 100 |
In vitro anticancer activity evaluation by SRB assay
The cell lines were grown in RPMI-1640 medium containing 10% fetal bovine serum and 2 mM L-glutamine. For the screening experiment cells were inoculated into 96 well microtiter plates in 90 μL at plating densities, depending on the doubling time of individual cell lines. After cell inoculation, the microtiter plates were incubated at 37 °C, 5% CO2, 95% air and 100% relative humidity for 24 h prior to addition of experimental compounds. After 24 h, one plate of each cell line was fixed in situ with TCA, to represent a measurement of the cell population for each cell line at the time of compound addition. Cisplatin and the test compounds were dissolved in dimethyl sulphoxide and stored frozen prior to use. The cytotoxicity assay was done against B16F10 (mouse melanoma), HeLa (human cervix cancer), A549 (human lung adenocarcinoma), SKOV3 (human ovarian adenocarcinoma) and MCF7 (human breast cancer) cell lines using a colorimetric SRB assay method. Solutions of different concentration of the complexes and cisplatin under test were added to the cell monolayer. Triplicate wells were prepared for each individual concentration. After compound addition, plates were incubated at standard conditions for 48 h and assay was terminated by the addition of cold trichloroacetic acid (TCA). Cells were fixed in situ by the gentle addition of 50 μL of cold 30% (w/v) TCA (final concentration, 10% TCA) and incubated for 60 minutes at 4 °C. Sulforhodamine B (SRB) solution (50 μL) at 0.4% (w/v) in 1% acetic acid was added to each of the wells, and plates were incubated for 20 minutes at room temperature. After staining, unbound dye was recovered and the residual dye was removed by washing five times with 1% acetic acid. Bound stain was subsequently eluted with 10 mM tris base (pH 10.5) for 5 min. The absorbance was read on an Elisa plate reader at a wavelength of 540 nm with 690 nm as reference wavelength. The results were expressed as the concentration at which there was 50% inhibition (IC50).Synthesis of ligand
Preparation of 2,5-di(1H-pyrazol-1-yl)-1,4-benzoquinone, C12H8N4O2
It was prepared by the reaction of pyrazole (2.5 g, 37 mmol) and p-benzoquinone (3.97 g, 37 mmol) in dry dioxane (15 mL). The mixture was heated under reflux for 6 h and the progress of the reaction was monitored by TLC. After completion of the reaction, the crude mixture was poured into ice-cold water and the resulting semi-solid that separated was extracted with ethyl acetate. The combined organic layers were dried over anhydrous sodium sulphate. It was then purified by column chromatography over silica gel using petroleum ether![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate (98
ethyl acetate (98![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) to yield 2,5-di(1H-pyrazol-1-yl)-1,4-benzoquinone. The product obtained was recrystallized from ethanol and compared with data reported.37
2) to yield 2,5-di(1H-pyrazol-1-yl)-1,4-benzoquinone. The product obtained was recrystallized from ethanol and compared with data reported.37
Yield: 42%. Mp: 206 °C. Anal. calcd for C12H8N4O2: C, 60.00; H, 3.36; N, 23.32%. Found: C, 60.55; H, 3.37; N, 23.29%. FT-IR (cm−1) in KBr: 1534 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O + C
O + C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1412 ν(C
C), 1412 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). UV-vis (DMSO), λmax: 257 (41
N). UV-vis (DMSO), λmax: 257 (41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 333), 311 (33
333), 311 (33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 403), nm (dm3 mol−1 cm−1). 1H NMR (CDCl3, ppm): 7.07 (s, 2H, quinone
403), nm (dm3 mol−1 cm−1). 1H NMR (CDCl3, ppm): 7.07 (s, 2H, quinone ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H; (Ha, Hb)), 7.67, 7.97 (2d, 4H,
C–H; (Ha, Hb)), 7.67, 7.97 (2d, 4H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H, J = 1.6 Hz; J = 2.4 Hz; (Hc, H′c), (He, H′e)), 6.45 (t, 2H, pyrazole
C–H, J = 1.6 Hz; J = 2.4 Hz; (Hc, H′c), (He, H′e)), 6.45 (t, 2H, pyrazole ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H, J = 2.0 Hz; J = 2.4 Hz; (Hd, H′d)). 13C NMR (CDCl3, ppm): 183.32 (ring C
C–H, J = 2.0 Hz; J = 2.4 Hz; (Hd, H′d)). 13C NMR (CDCl3, ppm): 183.32 (ring C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 156.01 (C–N), 139.20 (C
O), 156.01 (C–N), 139.20 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N) 107.79, 131.60, 142.15 (pyrazole).
C–N) 107.79, 131.60, 142.15 (pyrazole).
Synthesis of [Ru(L)Cl(CO)(PPh3)2] [complex 3]
A solution of [RuHCl(CO)(PPh3)3] (0.100 g, 0.105 mmol) in dry ethanol (15 mL) was refluxed for 15 minutes before adding the equimolar solution of the ligand 2,5-di(1H-pyrazol-1-yl)-1,4-benzoquinone (LH) (0.026 g, 0.105 mmol) in ethanol. The mixture was heated under reflux for 3 h, after which time the dark green solution was reduced in volume by rotary evaporation to approximately 10 mL. Then it was purified using silica gel column chromatography. Elutions with (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) pet ether–ethyl acetate gave the pure cyclometallated complex 3.
5) pet ether–ethyl acetate gave the pure cyclometallated complex 3.
Yield: 46%. Mp: 263 °C. Anal. calcd for C49H37N4O3P2Ru: C, 63.40; H, 4.02; N, 6.09%. Found: C, 63.49; H, 4.02; N, 6.10%. FT-IR (cm−1) in KBr: 1495 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O + C
O + C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1432 ν(C
C), 1432 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1936 ν(C
N), 1936 ν(C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 693, 1074, 1320 ν(PPh3). UV-vis (DMSO), λmax: 232 (95
O), 693, 1074, 1320 ν(PPh3). UV-vis (DMSO), λmax: 232 (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 740), 265 (65
740), 265 (65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), 302 (25
000), 302 (25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 820), nm (dm3 mol−1 cm−1) (intraligand transition); 449 (6670), (MLCT π*(LH) ← dπ(Ru)). 1H NMR (CDCl3, ppm): 6.62 (s, 1H, quinone
820), nm (dm3 mol−1 cm−1) (intraligand transition); 449 (6670), (MLCT π*(LH) ← dπ(Ru)). 1H NMR (CDCl3, ppm): 6.62 (s, 1H, quinone ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–Ha), 7.67, 7.61 (2d, 2H, free pyrazole –N
C–Ha), 7.67, 7.61 (2d, 2H, free pyrazole –N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H, J = 1.6 Hz; J = 2.0 Hz; (Hf, Hh)), 6.01 (t, 1H, free pyrazole
C–H, J = 1.6 Hz; J = 2.0 Hz; (Hf, Hh)), 6.01 (t, 1H, free pyrazole ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–Hg, J = 2.8 Hz), 8.77 (d, 1H, Ru–N
C–Hg, J = 2.8 Hz), 8.77 (d, 1H, Ru–N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–Hc, J = 2.0 Hz), 8.13 (d, 1H, bound prazole
C–Hc, J = 2.0 Hz), 8.13 (d, 1H, bound prazole ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–He, J = 2.4 Hz), 6.36 (t, 1H, bound pyrazole
C–He, J = 2.4 Hz), 6.36 (t, 1H, bound pyrazole ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–Hd, J = 2.4 Hz), 7.14–7.47 (m, 30H, aromatic PPh3). 13C NMR (CDCl3, ppm): 204.01 (C19
C–Hd, J = 2.4 Hz), 7.14–7.47 (m, 30H, aromatic PPh3). 13C NMR (CDCl3, ppm): 204.01 (C19![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 190.36 (C2–Ru), 172.79 (ring C1
O), 190.36 (C2–Ru), 172.79 (ring C1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 183.04 (ring O
O), 183.04 (ring O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C4–C–Ru), 140.42 (C
C4–C–Ru), 140.42 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C6–H), 141.03 (C5–N (free pyrazole)), 142.01 (C2–N (bound pyrazole)), 119.41 (C10
C6–H), 141.03 (C5–N (free pyrazole)), 142.01 (C2–N (bound pyrazole)), 119.41 (C10![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–C (free pyrazole)), 129.62 (C9
C–C (free pyrazole)), 129.62 (C9![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–C (bound pyrazole)), 108.01 (C11–C
C–C (bound pyrazole)), 108.01 (C11–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N (free pyrazole)), 108.33 (C8
N (free pyrazole)), 108.33 (C8![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–C (bound pyrazole)), 128.46 (C12
C–C (bound pyrazole)), 128.46 (C12![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–N (free pyrazole)), 140.80 (C7
N–N (free pyrazole)), 140.80 (C7![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–N (bound pyrazole)), 132.20, 132.86, 133.31, 133.40 (aromatic PPh3).
N–N (bound pyrazole)), 132.20, 132.86, 133.31, 133.40 (aromatic PPh3).
Synthesis of [Ru(L)Cl(CO)(AsPh3)2] [complex 4]
Treatment of 2,5-di(1H-pyrazol-1-yl)-1,4-benzoquinone (LH) (0.026 g, 0.106 mmol) with [RuHCl(CO)(AsPh3)3] (0.115 g, 0.106 mmol) under identical conditions to those used to prepare complex 3 (Scheme 1) afforded 4.Yield: 49%. Mp: 285 °C. Anal. calcd for C49H37N4O3As2Ru: C, 57.80; H, 3.66; N, 5.50. Found: C, 57.89; H, 3.66; N, 5.49. FT-IR (cm−1) in KBr: 1480 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O + C
O + C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1430 ν(C
C), 1430 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1924 ν(C
N), 1924 ν(C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 695, 1089, 1316 ν(AsPh3). UV-vis (DMSO), λmax: 236 (134
O), 695, 1089, 1316 ν(AsPh3). UV-vis (DMSO), λmax: 236 (134![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 710), 261 (117
710), 261 (117![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 342), 298 (61
342), 298 (61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 342), nm (dm3 mol−1 cm−1) (intraligand transition); 489 (8370), (MLCT π*(LH) ← dπ(Ru)). 1H NMR (CDCl3, ppm): 6.48 (s, 1H, quinone
342), nm (dm3 mol−1 cm−1) (intraligand transition); 489 (8370), (MLCT π*(LH) ← dπ(Ru)). 1H NMR (CDCl3, ppm): 6.48 (s, 1H, quinone ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–Ha), 7.64, 7.61 (2d, 2H, free pyrazole –N
C–Ha), 7.64, 7.61 (2d, 2H, free pyrazole –N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H, J = 1.6 Hz; J = 2.8 Hz; (Hf, Hh)), 6.05 (t, 1H, free pyrazole
C–H, J = 1.6 Hz; J = 2.8 Hz; (Hf, Hh)), 6.05 (t, 1H, free pyrazole ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–Hg, J = 2.8 Hz), 8.73 (d, 1H, Ru–N
C–Hg, J = 2.8 Hz), 8.73 (d, 1H, Ru–N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–Hc, J = 2.0), 8.14 (d, 1H, bound prazole
C–Hc, J = 2.0), 8.14 (d, 1H, bound prazole ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–He, J = 2.4 Hz), 6.30 (t, 1H, bound pyrazole
C–He, J = 2.4 Hz), 6.30 (t, 1H, bound pyrazole ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–Hd, J = 2.4 Hz), 7.13–7.24 (m, 30H, aromatic AsPh3). 13C NMR (CDCl3, ppm): 207.27 (C19
C–Hd, J = 2.4 Hz), 7.13–7.24 (m, 30H, aromatic AsPh3). 13C NMR (CDCl3, ppm): 207.27 (C19![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 193.12 (C2–Ru), 173.51 (ring C1
O), 193.12 (C2–Ru), 173.51 (ring C1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 181.75 (ring O
O), 181.75 (ring O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C4–C–Ru), 140.84 (C
C4–C–Ru), 140.84 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C6–H), 141.92 (C5–N (free pyrazole)), 143.27 (C2–N (bound pyrazole)), 121.15 (C10
C6–H), 141.92 (C5–N (free pyrazole)), 143.27 (C2–N (bound pyrazole)), 121.15 (C10![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–C (free pyrazole)), 128.54 (C9
C–C (free pyrazole)), 128.54 (C9![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–C (bound pyrazole)), 103.94 (C11–C
C–C (bound pyrazole)), 103.94 (C11–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N (free pyrazole)), 108.21 (C8
N (free pyrazole)), 108.21 (C8![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–C (bound pyrazole)), 127.90 (C12
C–C (bound pyrazole)), 127.90 (C12![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–N (free pyrazole)), 140.28 (C7
N–N (free pyrazole)), 140.28 (C7![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–N (bound pyrazole)), 133.07, 133.44, 133.91, 135.75 (aromatic AsPh3).
N–N (bound pyrazole)), 133.07, 133.44, 133.91, 135.75 (aromatic AsPh3).
Results and discussion
Synthesis and characterization of the complexes
Straightforward reactions of 2,5-di(1H-pyrazol-1-yl)-1,4-benzoquinone (LH) with [RuHCl(CO)(PPh3)3]/[RuHCl(CO)(AsPh3)3] gave new cyclometallated complexes of the type [RuLCl(CO)(EPh3)2] (where E = P/As), as depicted in Scheme 1. The complexes are diamagnetic which correspond to the bivalent state of ruthenium (low-spin d6, S = 0). The ligand was observed to undergo C–H activation at one of the ortho positions leading to the formation of five-member metallacycle. The complexes were stable to air and light, non-hygroscopic in nature and were remarkably soluble in chloroform, dichloromethane, DMF and DMSO and insoluble in water. The new complexes were characterized by elemental analysis, IR, 1H and 13C NMR spectroscopic techniques. The complexes were analytically pure as their microanalytical data confirmed the proposed molecular formula. It has been observed that a molecule of 2,5-di(1H-pyrazol-1-yl)-1,4-benzoquinone replaced a hydride ion and one of the triphenylphosphines/triphenylarsines from the precursor complexes. The solid state structure of both of the complexes (3 and 4) was determined by single crystal X-ray crystallographic studies. It revealed that LH behaved as monobasic bidentate donor.Spectroscopic studies
The IR spectrum of the free 2,5-di(1H-pyrazol-1-yl)-1,4-benzoquinone (LH) showed peaks of principal IR frequencies around 2855–3148, 1534 and 1412 cm−1 for ν(C–H), ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O + C
O + C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) and ν(C
C) and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) respectively. The band due to ν(C
N) respectively. The band due to ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O + C
O + C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) in the free ligand was found to be shifted towards lower wave numbers and the band due to ν(C
C) in the free ligand was found to be shifted towards lower wave numbers and the band due to ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) moved to higher wave numbers in the complexes. Thus, it is possible to assume that the ligand could give its coordination to ruthenium ion through O, N or even carbon. The appearance of new band around 1920–1936 cm−1 for terminal carbonyl vibrational responses in complexes 3 and 4 evidenced the complexation of precursor complexes with the ligand LH. The electronic spectrum of complexes 3 and 4 displayed absorption maxima at the range 232–489 nm. The intense absorptions at 232–236 nm and 261–265 nm is most likely due to a transition involving only ligand orbitals (π → π* and n → π*). The intense absorption observed in the visible region at 298–300 nm and at 449–489 nm are probably due to metal-to-ligand charge transfer transitions. These charge transfer transitions may be taking place from the t2 orbital of ruthenium (highest occupied molecular orbital) to the vacant π*(–C
N) moved to higher wave numbers in the complexes. Thus, it is possible to assume that the ligand could give its coordination to ruthenium ion through O, N or even carbon. The appearance of new band around 1920–1936 cm−1 for terminal carbonyl vibrational responses in complexes 3 and 4 evidenced the complexation of precursor complexes with the ligand LH. The electronic spectrum of complexes 3 and 4 displayed absorption maxima at the range 232–489 nm. The intense absorptions at 232–236 nm and 261–265 nm is most likely due to a transition involving only ligand orbitals (π → π* and n → π*). The intense absorption observed in the visible region at 298–300 nm and at 449–489 nm are probably due to metal-to-ligand charge transfer transitions. These charge transfer transitions may be taking place from the t2 orbital of ruthenium (highest occupied molecular orbital) to the vacant π*(–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–, –C
C–, –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N– or –C
N– or –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O–) orbital of the 2,5-di(1H-pyrazol-1-yl)-1,4-benzoquinone (lowest unoccupied molecular orbital) or to the higher energy vacant orbitals of other fragment of ligand. Complexation of ruthenium to the pyrazole nitrogen and to the quinone carbon rather than to the quinone oxygen was confirmed by NMR data (Experimental section). NMR spectra of the ligand and new complexes are given in Fig. S1–S5 (ESI†) (labeling scheme can found in Fig. 1).
O–) orbital of the 2,5-di(1H-pyrazol-1-yl)-1,4-benzoquinone (lowest unoccupied molecular orbital) or to the higher energy vacant orbitals of other fragment of ligand. Complexation of ruthenium to the pyrazole nitrogen and to the quinone carbon rather than to the quinone oxygen was confirmed by NMR data (Experimental section). NMR spectra of the ligand and new complexes are given in Fig. S1–S5 (ESI†) (labeling scheme can found in Fig. 1).
The 1H-NMR spectrum of the free ligand showed two protons singlet, a pair of two proton doublets and a two proton triplet assigned to (Ha, Hb), (Hc, H′c), (He, H′e) and (Hd, H′d) protons respectively. Complex 3 and 4 showed a sharp one proton singlet at δ 6.48 and 6.62 ppm which can be assigned to the Ha proton of the quinone ring. Generally complexation reduces the residual electron density on the ligands, which has a deshielding effect on the resonance field. Consistent with the above fact, the protons Hc, Hd, He of coordinated pyrazole resonated in down fields in comparison to free pyrazole protons Hf, Hg, Hh. A low field doublets at δ = 8.77 ppm (J = 2.0 Hz) for 3 and δ = 8.73 ppm (J = 2.0 Hz) and for 4 are assigned to the Hc proton that is nearer to the coordinated nitrogen atom. It is more down fielded than any other pyrazole protons of the ligand LH. Hf proton is observed as a doublet at δ 7.67 ppm (J = 1.6 Hz) for 3 and δ = 7.64 ppm (J = 1.6 Hz) for 4 respectively. Hd and Hg protons of complex 3 appeared as triplets at δ 6.36 ppm (J = 2.4 Hz) and δ 6.01 ppm (J = 2.8 Hz) respectively. However, in complex 4 they were appeared at δ 6.30 and δ 6.05 ppm. He and Hh protons resonated as doublets at δ 8.13 ppm (J = 2.4 Hz) and δ 7.61 ppm (J = 3.2 Hz). The aromatic protons of PPh3/AsPh3 in complex 3 and 4 were observed as multiplet around δ 7.13–7.47 ppm. In the 13C-NMR spectra of the complex 3 and 4, a highly downfield sharp singlet at δ 204.01 and 207.27 ppm has been assigned to the terminal carbonyl carbon C19. The next downfield singlet at δ 190.36 and 193.12 ppm is due to benzoquinone carbon C3 which is directly bonded to ruthenium. It is more down fielded than any other benzoquinone carbon atoms of LH ligand. C6 (sp2, low intensity band at δ 140.42 and 140.48 ppm respectively) is around 50 ppm up field when compared to that of C3. C1 and C4 carbons are appeared as singlets at δ 172.79 and 183.04 ppm respectively for 3 and for the same carbon atoms in 4 is at δ 173.51 ppm and 181.75 ppm respectively. C4 carbon is deshielded when compared to C1 because C4 is directly attached to C3, which is coordinated to ruthenium ion. A couple of singlets at δ 142.01 ppm and δ 141.03 ppm (for 3) and δ 143.27 ppm and δ 141.92 ppm (for 4) are due to the C2 and C5 carbons which are coupled to pyrazole nitrogens. C7 is resonated as singlet at δ 140.80 ppm and 140.28 ppm for 3 and 4 respectively. It is deshielded than any other pyrazole carbon atoms, because it is adjacent to nitrogen through which LH is bonded to ruthenium. Signals at δ 108.33 and δ 129.62 ppm for 3 and δ 108.21 ppm and δ 128.54 ppm for 4 are the resonances of bound pyrazole carbons C8 and C9 and signals at δ 128.46 ppm, δ 119.41 ppm and δ 108.01 ppm are due to C12, C10 and C11 of free pyrazole moiety respectively for complex 3. Whereas in complex 4 these resonances were observed at δ 127.90 ppm, δ 121.15 ppm and δ 103.94 ppm respectively. All carbon atoms of the six phenyl groups showed only 4 singlet signals in the range of 132.20–135.75 ppm. All the above points are in agreement with the interpretation of IR spectra suggesting bidentate coordination of the ligand to ruthenium and support the results of single crystal X-ray crystallographic studies.
X-ray crystallography
The molecular structures of 3 and 4 have been determined by single crystal X-ray analysis. The ORTEP drawings of 3 and 4 are displayed in Fig. 2 and 3 respectively along with their atom labeling scheme. The unit cell packing diagrams and intermolecular interactions of the complexes are given in Fig. S6 and S7 (ESI†). The summary of single crystal X-ray structures refinement is shown in Table 1. The selected geometrical parameters (inter atomic distances and angles) and hydrogen bond distances are given in Table 2. Single crystals of complexes 3 and 4 were obtained from slow evaporation of the mixture of dichloromethane, ethanol and ethyl acetate at room temperature as blue wedges. From the unit cell dimensions, it is clear that the crystal system of 3 and 4 is triclinic belonging to the space group P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) . In the new complexes 3 and 4, the coordination geometry around the ruthenium(II) ion is a slightly distorted octahedron C1N1(CO)Cl1E2, (E = P/As) where the ligand LH is coordinated to the metal by utilizing its pyrazole nitrogen and quinone carbon atom. A chloride and a carbonyl carbon complete the CN(CO)Cl square plane. As commonly observed for hexa coordinated complexes containing the {Ru(EPh3)2} unit, the two bulky triphenylphosphine/triphenylarsine molecules occupy the remaining two axial sites.38 The +2 oxidation state of the complexes is compensated by a chloride ion and a carbanion of benzoquinone ring. The ligand coordinated equatorially to ruthenium having the bite angle of C(2)–Ru(1)–N(1) = 77.40(6)° in complex 3 and 77.52(7)° in complex 4. This constrained bite angle has resulted in significant distortion in {CN(CO)ClE2} core from the ideal octahedral geometry, which is reflected in the twelve cis and three trans angles.
. In the new complexes 3 and 4, the coordination geometry around the ruthenium(II) ion is a slightly distorted octahedron C1N1(CO)Cl1E2, (E = P/As) where the ligand LH is coordinated to the metal by utilizing its pyrazole nitrogen and quinone carbon atom. A chloride and a carbonyl carbon complete the CN(CO)Cl square plane. As commonly observed for hexa coordinated complexes containing the {Ru(EPh3)2} unit, the two bulky triphenylphosphine/triphenylarsine molecules occupy the remaining two axial sites.38 The +2 oxidation state of the complexes is compensated by a chloride ion and a carbanion of benzoquinone ring. The ligand coordinated equatorially to ruthenium having the bite angle of C(2)–Ru(1)–N(1) = 77.40(6)° in complex 3 and 77.52(7)° in complex 4. This constrained bite angle has resulted in significant distortion in {CN(CO)ClE2} core from the ideal octahedral geometry, which is reflected in the twelve cis and three trans angles.
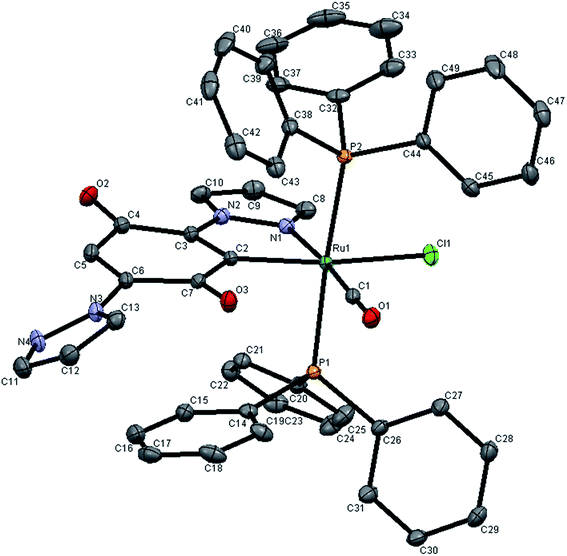 | ||
| Fig. 2 X-ray crystal structure with atom numbering scheme for 3 as thermal ellipsoids at 50% probability level. The hydrogen atoms have been omitted for clarity. | ||
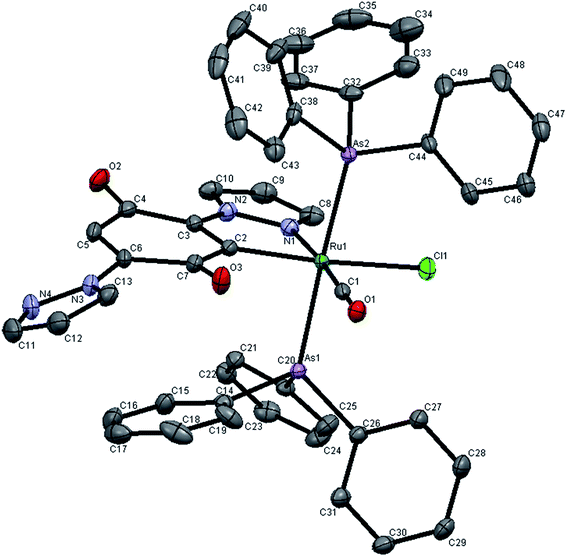 | ||
| Fig. 3 X-ray crystal structure with atom numbering scheme for 4 as thermal ellipsoids at 50% probability level. The hydrogen atoms have been omitted for clarity. | ||
| CCDC deposit number | 797114 | 797115 |
| Empirical formula | C50.23H40.69ClN4O3.62P2Ru | C49.77H42.31As2ClN4O3.385Ru |
| Formula weight | 956.62 | 1036.95 |
| Temperature/K | 90.0(5) | 90.0(5) |
| Crystal system | Triclinic | Triclinic |
| Space group | P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
| a/Å | 12.8966(10) | 12.9529(9) |
| b/Å | 13.2531(12) | 13.5239(10) |
| c/Å | 13.3050(14) | 13.2161(10) |
| α/° | 87.876(5) | 88.945(6) |
| β/° | 80.577(6) | 81.494(5) |
| γ/° | 70.558(6) | 69.810(5) |
| Volume/Å3 | 2115.1(3) | 2147.6(3) |
| Z | 2 | 2 |
| ρcalc/mg mm−3 | 1.502 | 1.604 |
| m/mm−1 | 0.561 | 2.006 |
| F(000) | 980.0 | 1046.0 |
| Crystal size/mm3 | 0.20 × 0.18 × 0.07 | 0.23 × 0.22 × 0.10 |
| 2θ range for data collection | 5.4 to 65° | 5.8 to 66.2° |
| Index ranges | −19 ≤ h ≤ 19, −20 ≤ k ≤ 20, −20 ≤ l ≤ 20 | −19 ≤ h ≤ 19, −20 ≤ k ≤ 20, −20 ≤ l ≤ 20 |
| Reflections collected | 62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 665 665 |
61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 539 539 |
| Independent reflections | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 231 [R(int) = 0.044] 231 [R(int) = 0.044] |
16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 353 [R(int) = 0.036] 353 [R(int) = 0.036] |
| Data/restraints/parameters | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 231/0/542 231/0/542 |
16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 353/0/541 353/0/541 |
| Goodness-of-fit on F2 | 1.069 | 1.037 |
| Final R indexes [I ≥ 2σ(I)] | R1 = 0.0390, wR2 = 0.0930 | R1 = 0.0360, wR2 = 0.0840 |
| Final R indexes [all data] | R1 = 0.0570, wR2 = 0.0980 | R1 = 0.0620, wR2 = 0.0910 |
| Largest diff. peak/hole/e Å−3 | 0.67/−0.89 | 1.47/−0.96 |
| Inter atomic distances (Å) | |||
|---|---|---|---|
| Complex 3 | Complex 4 | ||
| Ru(1)–N(1) | 2.126(1) | Ru(1)–Cl(1) | 2.4930(6) |
| Ru(1)–C(2) | 2.029(2) | Ru(1)–C(1) | 1.853(2) |
| Ru(1)–C(1) | 1.857(1) | Ru(1)–C(2) | 2.024(2) |
| Ru(1)–Cl(1) | 2.4870(5) | Ru(1)–N(1) | 2.120(1) |
| Ru(1)–P(1) | 2.3941(5) | Ru(1)–As(1) | 2.4645(3) |
| Ru(1)–P(2) | 2.4045(6) | Ru(1)–As(2) | 2.4759(3) |
| Bond angles (°) | |||
|---|---|---|---|
| Complex 3 | Complex 4 | ||
| C(2)–Ru(1)–P(1) | 89.78(5) | C(1)–Ru(1)–Cl(1) | 98.99(7) |
| C(2)–Ru(1)–C(1) | 97.61(7) | C(1)–Ru(1)–C(2) | 97.53(8) |
| C(2)–Ru(1)–Cl(1) | 161.93(5) | C(1)–Ru(1)–N(1) | 175.00(8) |
| C(2)–Ru(1)–N(1) | 77.40(6) | C(1)–Ru(1)–As(1) | 86.91(6) |
| C(2)–Ru(1)–P(2) | 92.53(5) | C(1)–Ru(1)–As(2) | 89.48(6) |
| C(1)–Ru(1)–Cl(1) | 100.46(6) | C(2)–Ru(1)–As(1) | 88.83(6) |
| C(1)–Ru(1)–P(1) | 85.75(6) | C(2)–Ru(1)–As(2) | 92.65(6) |
| C(1)–Ru(1)–N(1) | 174.91(7) | C(2)–Ru(1)–N(1) | 77.52(7) |
| C(1)–Ru(1)–P(2) | 89.32(6) | C(2)–Ru(1)–Cl(1) | 163.45(6) |
| N(1)–Ru(1)–Cl(1) | 84.53(4) | N(1)–Ru(1)–Cl(1) | 85.95(5) |
| N(1)–Ru(1)–P(1) | 93.17(4) | N(1)–Ru(1)–As(1) | 92.29(5) |
| N(1)–Ru(1)–P(2) | 91.89(4) | N(1)–Ru(1)–As(2) | 91.40(5) |
| Cl(1)–Ru(1)–P(1) | 91.11(2) | Cl(1)–Ru(1)–As(1) | 91.10(1) |
| Cl(1)–Ru(1)–P(2) | 88.14(2) | Cl(1)–Ru(1)–As(2) | 88.46(1) |
| P(1)–Ru(1)–P′(2) | 174.79(2) | As(1)–Ru(1)–As′(2) | 176.24(1) |
| Inter and intra molecular interactions | ||
|---|---|---|
| Complex 3 | O3⋯O1 (intra) | O3⋯O1 (inter) |
| 2.988(2) | 2.992(2) | |
| Complex 4 | O3⋯O1 | |
| 2.974(2) | ||
The cis bond angles spread over the range 84.53(4)–100.46(6)°, while the trans bond angles are within the range 161.93(5)–176.24(1)°. The carbonyl group occupies a site trans to the N1 (pyrazole ring nitrogen from LH, N1–Ru1–C1, 174.91(7) Å in 3175.00(8) Å in 4). This may be a consequence of strong Ru(II) → CO back donation as indicated by the short Ru–C bond (1.857(1) Å) and low CO stretching frequency (1936 cm−1), which prefers σ or weak π donor groups occupying the site opposite to CO to favor the d–π* back donation. Though the triphenylphosphine/triphenylarsine ligands usually prefer to occupy mutually cis position for better π-interaction,39 in this complex the presence of CO, a stronger π-acidic ligand, might have forced the bulky triphenylphosphine/triphenylarsine ligands to take trans position for steric reasons. The C(2)–Ru(1)–Cl(1) bond angle is 161.93(5) Å and 163.45(6) Å in complexes 3 and 4 respectively showing that Cl atom lies trans to ring carbon. From the covalent radii values, the Ru(II)–C(sp2) length is estimated to be 2.06 Å (ref. 40) and the observed values extent the range 1.96–2.16 Å.41 In the new complexes, the Ru1–C2 distance is 2.024–2.029 Å, which is identical to that of the other structurally characterized cyclometallated ruthenium(II) complexes.42 The Ru1–C1 bond distance (1.857 Å in complex 3 and 1.853 Å in complex 4) is in accordance with those found in another ruthenium(II) complex.43 The Ru–C2 (sp2) bond is 0.17 Å longer than the Ru1–C1 (sp) distance in complexes 3 and 4, due to Ru–CO back bonding. Among six Ru-ligand distances, the Ru1–Cl distance of 2.487 Å is significantly longer than other bond distances. This lengthening of the Ru–Cl bond distance can be explained by the trans influence of the soft aryl carbon atom which is present trans to the chloride. The Ru–P, Ru–As, Ru–O, Ru–N and Ru–Cl bond lengths found in the complexes agree well with those reported for similar ruthenium complexes having same coordination sphere.43–46 The absence of any solvent of crystallization in the crystal lattice of the new complexes indicates possible existence of non-covalent interaction(s) between the individual complex molecules. A closer look at the packing pattern of the crystal revealed that the carbonyl oxygen in each complex molecule exhibited inter and intra molecular interactions with benzoquinone oxygen atom.
DNA binding studies
Since DNA is an important cellular receptor, interaction with DNA, an elementary mechanism of action responsible for anticancer activity which has been suggested for some of the ruthenium complexes. Thus, the mode and propensity for binding of the ruthenium(II) precursor complexes 1, 2 and their corresponding new ruthenium(II) complexes 3, 4 to HS-DNA were studied with the aid of electronic absorption and luminescence quenching techniques. The ligand LH was also interacted with DNA to find out the impact of chelation on binding with biomolecules.Electronic absorption titration
The application of electronic absorption spectroscopy is one of the simplest and most useful techniques used to study the DNA-binding properties of the metal complexes. In the UV-vis spectra of the ligand and complexes, absorption bands were observed at 230 and 270 nm. In addition to these bands, the new complexes 3 and 4 showed additional low intense MLCT bands at 449 and 489 nm respectively. In order to study the binding of the test compounds (LH, ruthenium(II) starting materials 1, 2 and their corresponding new ruthenium(II) organometallic complexes 3, 4) with DNA, the change in the electronic spectrum was observed by titrating increasing concentrations of HS-DNA (0–50 μM). As the concentration of DNA was increased, the intensity of the absorption bands at 257, 267, 259, 265 and 261 nm corresponding to LH and complexes 1–4 respectively were affected by resulting hyperchromism with red shifts in the IL (intra-ligand) bands. The spectral changes of LH and complexes 1–4 upon addition of DNA are shown in Fig. 4. The observed spectroscopic variation i.e. hyperchromic effect with red shifts suggested that the test compounds bind to HS-DNA by external contact, possibly due to electrostatic binding.47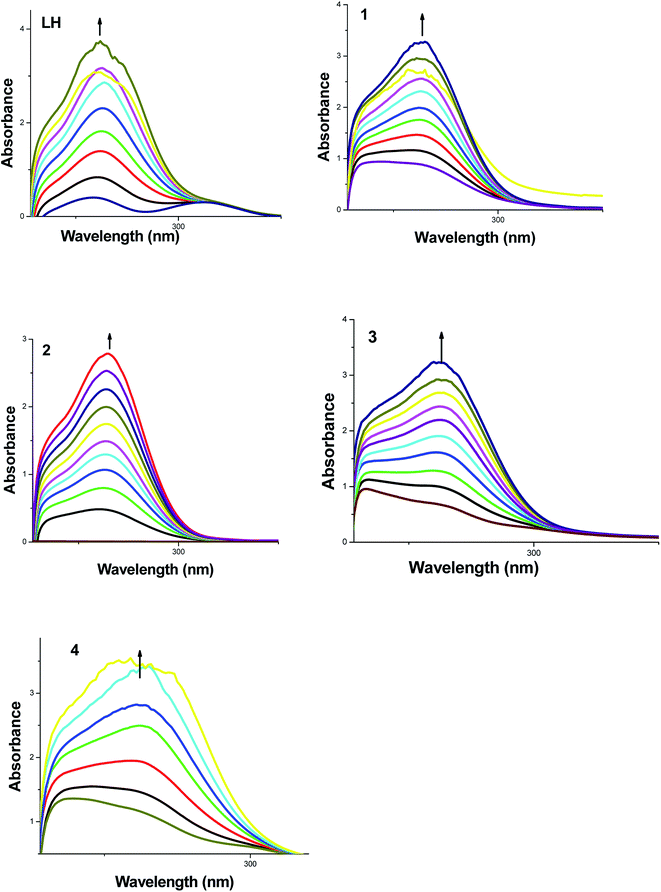 | ||
| Fig. 4 Absorption titration spectra of fixed concentration (10 μM) of LH and 1–4 with increasing concentrations (0–50 μM) of HS-DNA (phosphate buffer, pH 7). | ||
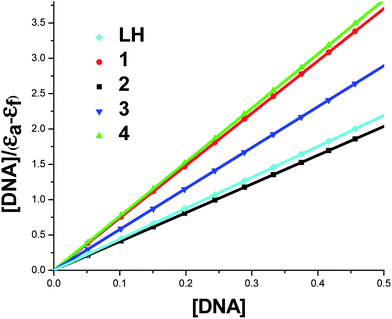
To compare quantitatively the binding affinity of the test compounds to DNA, the intrinsic binding constants Kb of the compounds with DNA were calculated by monitoring the changes in absorbance around 258–267 nm with increasing concentration of DNA using the following equation.
| [DNA]/[εa − εf] = [DNA]/[εb − εf] + 1/Kb[εb − εf] | (1) |
Wherein the present case, [DNA] is the concentration of DNA in base pairs, εa, εf, and εb are corresponding respectively to the extinction coefficient for each addition of DNA to the test compound, the extinction coefficient of free compound and the extinction coefficient of the test compound in the fully bound form. Kb is the equilibrium binding constant of the test compound binding to HS-DNA. Each set of data, when fitted to the above equation generate a straight line with a slope of 1/(εb − εf) and a y-intercept of 1/Kb (εb − εf) and Kb was determined from the ratio of the slope to intercept (Table 3 and Fig. 5).
| System | Kb (M−1) |
|---|---|
| HS-DNA + LH | 1.74 × 105 |
| HS-DNA + 1 | 1.21 × 104 |
| HS-DNA + 2 | 1.08 × 104 |
| HS-DNA + 3 | 2.18 × 106 |
| HS-DNA + 4 | 6.95 × 105 |
From the binding constant values (Table 3), it is inferred that the new organometallic complexes 3, 4 showed better binding affinity than the ligand LH and their parent complexes 1 and 2. Complex 3 exhibited better DNA binding than complex 4 and this may be due to the relatively small size of phosphine as compare with arsine.
Competitive binding studies with ethidium bromide
Though absorption spectral studies point out the electrostatic binding of the test compounds with DNA, it has to be reconfirmed by other methods and hence, steady-state competitive binding experiments using compounds LH, 1, 2, 3 and 4 as quenchers were undertaken to get final proof for the mode of binding of the compounds to DNA. For that ethidium bromide (EB) was employed as a fluorescent probe. EB is a classical intercalator exhibits a dramatic enhancement of its emission intensity when intercalated to DNA. This enhanced fluorescence of the EB–DNA complex can be quenched by the addition of a second DNA binder which is either an intercalator or a groove binder and such a decrease in the emission intensity of the pre-treated EB is the basis of the fluorescence intercalator displacement assay. In our experiment, the EB–DNA system exhibits a strong emission band at wavelength around 620 nm. Emission spectra of EB bound to HS-DNA in the absence and presence of each compound have been recorded for [EB] = 10 μM, [DNA] = 10 μM for increasing amounts of each compound (0–50 μM). Upon the addition of LH and complexes 1–4, the fluorescence emission of the EB–DNA system was enhanced rather than quenched in the emission maximum shown in Fig. S8 (ESI†), indicating that they could not displace EB from the EB–DNA complex. This observation also indicates that the compounds bind weakly to the DNA probably by electrostatic interaction which is supported by the absorption titration experiment.BSA binding studies
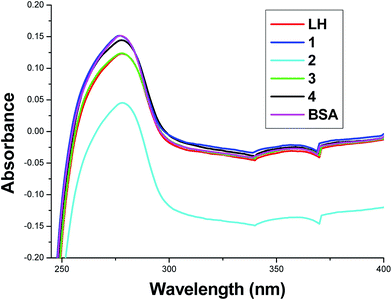
Since the mode of action of ruthenium(II)-based metallo drugs in chemotherapy, still a subject of debate. The investigation of interaction between metabolism products such as protein and ruthenium complexes is essential for a better understanding of their pharmacological profile. It has also been proved that organometallic ruthenium complexes are reactive towards a number of proteins and play a significant role in their biological activity48 and hence, investigations on the interactions of our compounds with protein were undertaken. Among the proteins, since bovine serum albumin is one of the most extensively studied proteins, particularly because of its medical importance, stability and structural homology with human serum albumin (HSA) and thus our model protein. The nature of interaction of our compounds (LH and complexes 1–4) with BSA has been explored by means of UV-vis and fluorescence spectroscopy.UV absorption spectra of BSA in the presence of compounds
UV-vis spectra of BSA in the absence and presence of the test compounds are shown in Fig. 6. It is seen from the figure that a progressive decrease in the absorbance intensity of BSA was observed with the addition of these test compounds and there was a small blue shift of 1 nm for 1 and red shift of about 1 nm for the compounds LH, 2, 3 and 4 respectively. This result suggested not only the complex formation between BSA and the test compounds but also confirms a static quenching process because the dynamic quenching mechanism does not change the electronic absorption spectrum of the compounds.49Fluorescence quenching studies of BSA
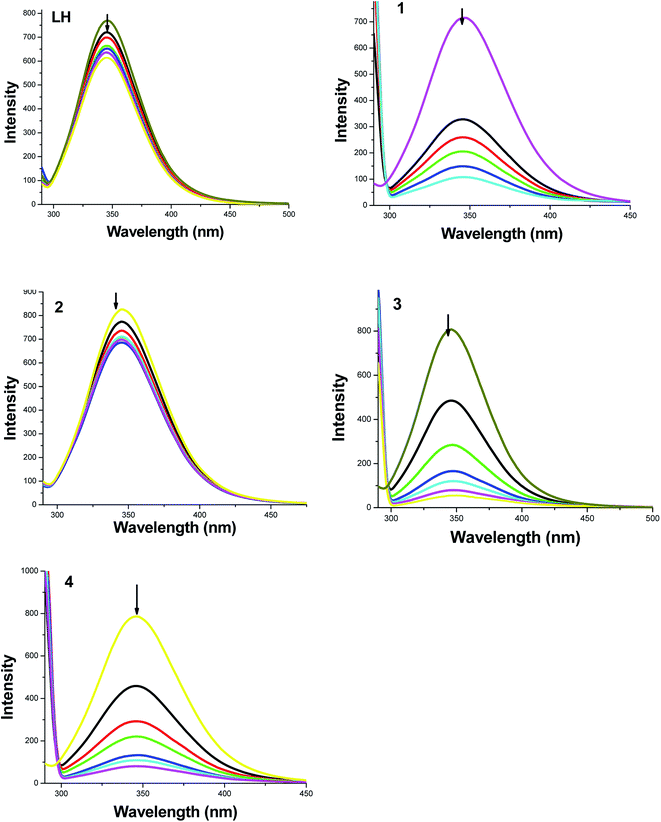
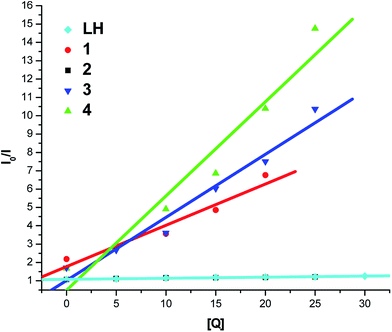
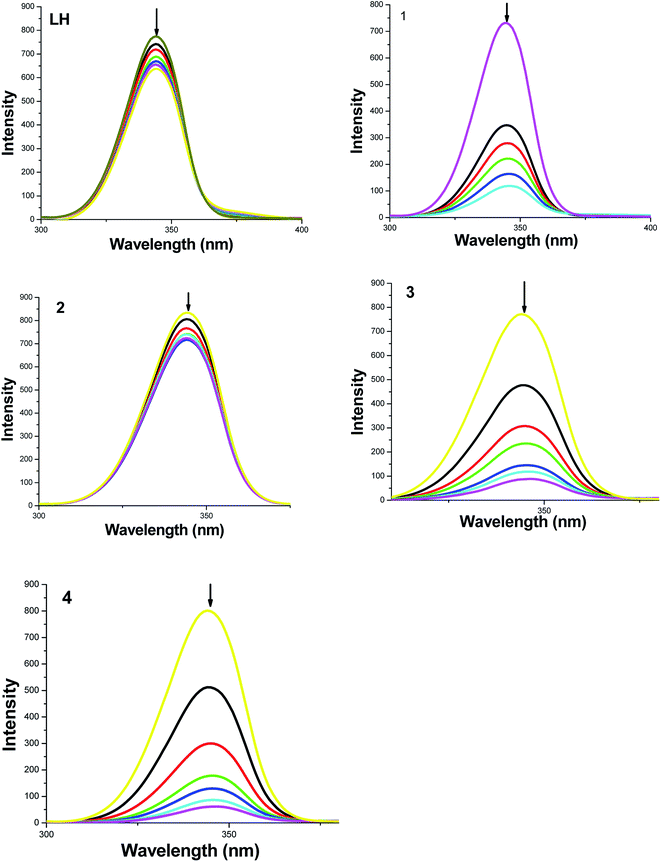
To get more information on the binding of the compounds with BSA, fluorescence spectrum of BSA was studied in the presence of the test compounds. BSA contains three fluorophores, namely, tryptophan, tyrosine and phenylalanine, the intrinsic fluorescence of BSA is mainly due to tryptophan alone50 and changes in the emission spectra of tryptophan are common in response to protein conformational transitions, subunit associations, substrate binding or denaturation. Therefore, the intrinsic fluorescence of BSA can provide considerable information on their structure and dynamics and is often employed in the study of protein folding and association reactions. Hence, the interaction of BSA with our ligand LH and complexes (1–4) was studied by fluorescence measurement at room temperature and from which the binding constants of the complexes were calculated. In a typical experiment, a solution of BSA (1 μM) was titrated with various concentrations of the complexes (0–35 μM). Fluorescence spectra were recorded in the range of 290–500 nm upon excitation at 280 nm. The changes observed on the fluorescence emission spectra of a solution of BSA on the addition of increasing amounts of test compounds are shown in Fig. 7. Addition of compounds to BSA produced a dramatic modification on the emission profile. The fluorescence of BSA was quenched effectively around 345 nm accompanied with the blue shift in emission maximum of LH, complexes 3, 4 and with the red shift in starting complexes 1, 2 and this may be due to the binding of compounds with the active site in BSA.51To study the quenching process further, fluorescence quenching data were analyzed with the Stern–Volmer eqn (2) and Scatchard eqn (3). The ratio of the fluorescence intensity in the absence of (I0) and in the presence of (I) the quencher is related to the concentration of the quencher [Q] by a coefficient KSV.
| I0/I = 1 + KSV[Q] | (2) |
KSV value obtained from the plot of I0/I vs. [Q] was found to be 5.87 × 103 M−1, 1.39 × 105 M−1, 5.10 × 103 M−1, 5.14 × 105 M−1 and 3.44 × 105 M−1 corresponding to LH and complexes 1–4 respectively. The observed linearity in the plots (Fig. 8 and Table 4) indicated the ability of the test compounds to quench the emission intensity of BSA. From KSV values, it is seen that the complexes 3 and 4 exhibited strong protein-binding ability with enhanced hydrophobicity than LH and their corresponding starting materials 1 and 2.
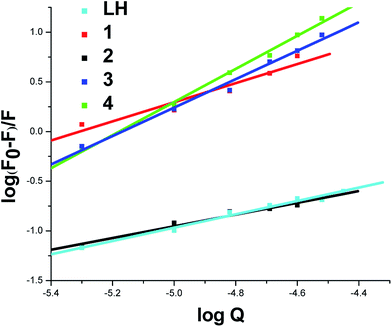
| System | KSV × M−1 | K × M−1 | n |
|---|---|---|---|
| BSA + LH | 5.87 × 103 | 2.44 × 104 | 0.67 |
| BSA + 1 | 1.39 × 105 | 1.26 × 105 | 0.96 |
| BSA + 2 | 5.10 × 103 | 1.00 × 105 | 0.59 |
| BSA + 3 | 5.14 × 105 | 4.24 × 106 | 1.06 |
| BSA + 4 | 3.44 × 105 | 2.47 × 106 | 1.43 |
For the static quenching, when small molecules bind independently to a set of equivalent sites on a macromolecule, the following Scatchard eqn (3) can be used to determine the binding constant and the number of binding sites.
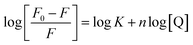 | (3) |
| IC50a (μM) | |||||
|---|---|---|---|---|---|
| Compounds | DPPH˙ | OH˙ | NO˙ | O2−˙ | H2O2 |
| a Fifty percent inhibitory concentration of the test compounds against free radicals. | |||||
| LH | 208.49 ± 0.7 | 122.52 ± 2.3 | 165.26 ± 1.9 | 181.1 ± 0.8 | 223.8 ± 0.5 |
| 1 | 199.7 ± 3.4 | 234.9 ± 1.5 | 212.2 ± 4.8 | 267.4 ± 3.6 | 288.5 ± 3.2 |
| 2 | 243.5 ± 4.5 | 289.8 ± 5.8 | 256.3 ± 5.9 | 294.2 ± 4.8 | 302.2 ± 2.7 |
| 3 | 21.27 ± 2.8 | 50.65 ± 6.1 | 65.96 ± 5.6 | 33.75 ± 1.3 | 157.1 ± 0.9 |
| 4 | 19.40 ± 1.1 | 57.88 ± 4.6 | 71.00 ± 0.9 | 47.06 ± 0.4 | 169.4 ± 1.5 |
| Vitamin C | 147.6 ± 4.2 | 232.8 ± 1.9 | 215.8 ± 2.7 | 221.4 ± 1.2 | 238.5 ± 3.6 |
| BHT | 86.2 ± 1.8 | 163.4 ± 0.7 | 154.3 ± 2.4 | 131.6 ± 1.5 | 149.8 ± 4.3 |
Synchronous fluorescence spectra
After having obtained the binding constant and number of binding sites of the test compounds with BSA, it is essential to know about the conformational change of the protein molecular environment in the vicinity of the fluorophore functional groups.52 This can be derived from synchronous fluorescence spectral studies and hence, the synchronous fluorescence spectra of BSA with the compounds were carried out. According to Miller,53 in synchronous fluorescence spectroscopy, the difference between excitation and emission wavelength (Δλ = λemi − λexc) reflects the spectra of a different nature of chromophores. If the Δλ value is 15 nm, the synchronous fluorescence of BSA is characteristic of tyrosine residue, whereas a larger Δλ value of 60 nm is characteristic of tryptophan.54 The variation in the tryptophan emission is the consequence of the protein conformational changes. Therefore, to explore the structural change of BSA, we measured synchronous fluorescence spectra of BSA with various concentration of LH and 1–4 at Δλ = 15 nm and Δλ = 60 nm and the changes are shown in Fig. S9 (ESI†) and Fig. 10 respectively.Results showed that the fluorescence of tyrosine residue was hyperchromic and that of tryptophan residue was hypochromic. However, the fluorescence intensities of tryptophan residues changed significantly with red shift by addition of increasing amount of the compounds especially in case of the new complexes 3 and 4. It indicates that the hydrophobicity of microenvironment around tryptophan residues decreases in the presence of the added compounds55 and that the complexes bind to active site in the protein, which makes them potential molecules for biological applications. This result has prompted us to explore more on the anticancer activities of the new organometallic ruthenium(II) complexes.
Antioxidant activity
Free radicals that are generated in many bioorganic redox processes may induce oxidative damage in various components of the body (lipids, DNA and proteins) and have been implicated in aging and a number of life-limiting chronic diseases such as cancer, hypertension, parkinson disease, alzheimer, cardiac infarction, atherosclerosis, rheumatism, cataracts etc.56,57 Efforts to counteract the damage caused by these species are gaining acceptance as a basis for novel therapeutic approaches and the field of preventive medicine is experiencing an upsurge of interest in medically useful antioxidants. Since the experiments conducted so far revealed that the complexes exhibit reasonable DNA and protein binding affinity, it is considered worthwhile to study their ability to quench the free radicals and antioxidant properties. Further, the in vitro antioxidant properties of ruthenium complexes have attracted a lot of interest, but the radical scavenging activity is limited to hydroxyl radical.58 Hence, we carried out experiments to investigate the free radical scavenging ability of the new ruthenium complexes against a group of free radicals. The ligand and the metallic precursors were also subjected to radical scavenging experiments. The IC50 value of all test compounds (Table 5) obtained from different types of assay experiments strongly supports that the complexes presented in this work possess excellent antioxidant activities, which are better than that of the standard antioxidants vitamin C and BHT.The new organometallic complexes 3 and 4 having similar structure showed almost comparable antioxidant activities. The next higher antioxidant activity was possessed by the free ligand LH and the precursor complexes 1 and 2 were observed to be less effective on the free radicals. The new ruthenium(II) complexes including the precursor complexes exhibited greater scavenging effect on stable free radical DPPH (IC50(4) = 19.40 ± 1.1 μM) and lower effect towards hydrogen peroxide (IC50(4) = 169.4 ± 1.5 μM). The DPPH and superoxide anion radical scavenging ability of the new complexes are better than that of those reported by us for other similar ruthenium(II) complexes38,59 and lower OH radical scavenging effect than other previously reported ruthenium complexes.58a,b,d It is to be noted that no significant radical scavenging activities were observed in all the experiments carried out with ruthenium precursor complexes and ligands under the same experimental conditions. From the above results, it is clear that the scavenging effects of the precursor complexes and free ligand is significantly less when compared to their corresponding ruthenium(II) complexes, which is mainly due to the chelation of the organic ligand with the ruthenium(II) ion.
Anticancer activity studies
Since the results of antioxidant activity experiments revealed that the new complexes exhibit significant radical scavenging activity, we interested to determine the cytotoxic activity of the new organometallic complexes because nowadays it has been strongly suspected that cancer may be one of those degenerative diseases induced by free radicals and reactive oxygen species. Hence, the antiproliferative activity of the new complexes 3 and 4 were tested and to stresses the influence of chelation on cytotoxic activity the ligand LH and ruthenium(II) precursor complexes 1 and 2 were also tested in a panel of cancer cell lines including B16F10 (mouse melanoma), A549 (human lung adenocarcinoma), SKOV3 (human ovarian adenocarcinoma), MCF7 (human breast cancer) and HeLa (human cervix cancer) by means of SRB assay. The effects of the test compounds on the viability of these cells were evaluated after an exposure period of 48 h. The cancer cells were treated with different concentrations of the test compounds. The test compounds were dissolved in DMSO and the blank samples containing same amount of DMSO were taken as controls to find out the activity of the solvent in the cytotoxicity experiment. In parallel, the influence of well established anticancer drug, cisplatin has also been assayed as a positive control. It is to be noted that the ruthenium precursor complexes 1, 2 and the ligand did not produce any noteworthy inhibition of cell growth even after >100 μM of concentration on cancer cells implying that the ligand and ruthenium precursor complexes are virtually devoid of antiproliferative properties and the chelation of the ligand with the ruthenium ion being the responsible factor for the observed cytotoxic properties of the new organometallic complexes. The IC50 values for two new ruthenium complexes 3, 4 and cisplatin for selected cell lines are shown in Table 6.| Complexes | IC50a (μM) | ||||
|---|---|---|---|---|---|
| B16F10 | HeLa | A549 | MCF7 | SKOV3 | |
| a Fifty percent inhibitory concentration after exposure for 48 h in the SRB assay. | |||||
| 3 | 43.25 ± 1.03 | 12.60 ± 2.93 | 48.90 ± 1.73 | 19.10 ± 3.93 | 30.30 ± 0.49 |
| 4 | 47.10 ± 1.58 | 16.90 ± 2.12 | 55.30 ± 4.11 | 40.97 ± 3.55 | 36.20 ± 1.98 |
| Cisplatin | 5.54 ± 0.01 | 16.20 ± 0.7 | 5.62 ± 0.28 | 7.73 ± 0.11 | 9.61 ± 0.52 |
As shown in Table 6, the new ruthenium(II) organometallic complexes 3 and 4 showed moderate to potent anticancer activities against the tested cells, with IC50 values ranging from 12.60 ± 2.93 μM to 55.30 ± 4.11 μM. Out of the five cancer cell lines chosen to examine, the activity corresponding to inhibition strength of the tested organometallic complexes was most against HeLa cells and least in A549 cells. In general 3 and 4 exhibited comparable cytotoxic properties except in the case of MCF7 cell inhibition, where the inhibitory activity of 4 was found to be two times lower than 3. Complex 3 (IC50 = 12.60 ± 2.93 μM) outperforming the inhibitory activity of cisplatin (IC50 = 16.20 ± 0.70 μM) against HeLa cells and 4 (IC50 = 16.90 ± 2.12 μM) displayed equipotent activity. The observed cytotoxic effects by the new ruthenium complexes toward the HeLa and MCF-7 cell lines are much better than those previously reported for other Ru complexes.58a,b,d,60 On the whole, there is no substantial variation in the cytotoxic activities when triphenylphosphine is replaced by triphenylarsine in the complexes which is consistent with the trends of their radical scavenging abilities. Though the new ruthenium complexes display the contemporary absence of direct cell cytotoxicity (in vitro), further studies are needed to assess their antiproliferative activity in vivo and to elucidate the actual mechanism of the anti-tumor activity.
Conclusions
Two new organo ruthenium(II) complexes have been prepared with the main emphasis on evaluation of chelation on anticancer activity by comparing the activities with their respective precursor complexes and the ligand. The new complexes were characterized by elemental analysis and various spectroscopic techniques. The monobasic bidentate coordination of the ligand, the metal mediated C–H activation along with the molecular structure of both the new complexes 3 and 4 were authenticated by single crystal X-ray diffraction studies. Such C–H activation is quite surprising in the case of quinone type ligands even in the presence of electron rich donor (oxygen atoms). While comparing the binding ability of chelator LH, metallic precursors 1, 2 and new ruthenium chelates 3 and 4 with HS-DNA/BSA, the new ruthenium complexes exhibiting better binding ability with the biomolecules than the ligand and their parent complexes. The results obtained from various antioxidant assays showed their significant efficacy in scavenging the radicals and the assumed patterns of activity increased in the following order 1, 2 < LH < 3–4. The cytotoxicity of the compounds against a panel of cancer cells demonstrated that the new complexes 3 and 4 showed promising tumor cell growth inhibiting activity and outperforming than 1, 2, LH against all cells and cisplatin in the case of HeLa cells. The above said pharmacological investigations support the fact that there exists a strong influence of chelation on anticancer activity. The significant outcome of the present investigation regarding the abilities of new ruthenium organometallic complexes towards various biological evaluations is that chelation notably increased the interaction with biomolecules such as DNA/BSA resulted in exhibiting excellent radical scavenging ability and considerable cytotoxicity.Acknowledgements
Department of Science and Technology (SR/FT/CS-32-/2011 dated 04.05.2012) and Council of Science and Industrial Research (01(2553)/12/EMR-II dated 03.04.2012) New Delhi, India are greatly acknowledged for their financial support.References
- (a) M. N. Ibrahim and E. S. Sharif, E-J. Chem., 2007, 4, 531 CrossRef CAS; (b) Z. X. Yan, S. H. Li, Q. Liux and L. Tange, Polyhedron, 2007, 26, 3743 CrossRef.
- M. Revanasiddappa, T. Suresh, K. Syed and S. D. Angadi, E-J. Chem., 2008, 5, 395 CrossRef CAS.
- E. Labisbal, K. D. Haslow, A. Sousapedrares, J. Valdes-Martnez, S. Hern and D. X. West, Polyhedron, 2003, 22, 2831 CrossRef CAS.
- R. Morphy and Z. Rankovic, J. Med. Chem., 2005, 48, 6523 CrossRef CAS PubMed.
- J. M. Brown and A. J. Giaccia, Cancer Res., 1998, 58, 1408 CAS.
- G. R. Fisher, J. Donis and P. L. Gutierrez, Biochem. Pharmacol., 1992, 44, 1625 CrossRef CAS PubMed.
- J. Cummings, V. J. Spanswick, M. Tomasz and J. F. Smyth, Biochem. Pharmacol., 1998, 56, 405 CrossRef CAS PubMed.
- L. Ernster, Chem. Scr., 1987, 27A, 1 CAS.
- W. Ma, H. Zhou, Y. L. Ying, D. W. Li, G. R. Chen, Y. T. Long and H. Y. Chen, Tetrahedron, 2011, 67, 5990 CrossRef CAS.
- N. M. Ruvalcaba, G. Cuevas, I. Gonzalez and M. A. Martinez, J. Org. Chem., 2002, 67, 3673 CrossRef.
- Y. Izumi, H. Sawada, N. Sakka, N. Yamamotto, T. Kume, H. Katsuki, S. Shimohama and A. Akaike, J. Neurosci. Res., 2005, 79, 849 CrossRef CAS PubMed.
- R. Meganathan, Vitam. Horm., 2001, 61, 173 CAS.
- S. L. Planz, H. H. Sedlacek and S. Pleschka, PCT Int. Appl., WO2004085682, 2004.
- E. M. Perchellet, M. M. Ward, A. L. Skaltsounis, I. K. Kostakis, N. Pouli, P. Marakos and J. H. Perchellet, Anticancer Res., 2006, 26, 2791–2804 CAS.
- B. Insuasty, A. Tigreros, F. Orozco, J. Quiroga, R. Abonia, A. Sanchez and J. Cobo, Bioorg. Med. Chem., 2010, 18, 4965 CrossRef CAS PubMed.
- M. Labbozzetta, R. Baruchello, P. Marchetti, M. C. Gueli, P. Poma, M. Notarbartolo, D. Simoni and D. N. Alessandro, Chem.-Biol. Interact., 2009, 181, 29 CrossRef CAS PubMed.
- P. Pevarello, M. G. Brasca, R. Amici, P. Orsini, G. Traquandi, L. Corti, C. Piutti, P. Sansonna, M. Villa, B. S. Pierce, M. Pulici, P. Giordano, K. Martina, E. L. Fritzen, R. A. Nugent, E. Casale, A. Cameron, M. Ciomei, F. Roletto, A. Isacchi, G. P. Fogliatto, E. Pesenti, W. Pastori, A. Marsiglio, K. L. Leach, P. M. Clare, F. Fiorentini, M. Varasi, A. Vulpetti and M. A. Warpehoski, J. Med. Chem., 2004, 47, 3367 CrossRef CAS PubMed.
- M. Yoshimoto, H. Miyazawa, H. Nakao, K. Shinkai and M. Arakawa, J. Med. Chem., 1979, 22, 491 CrossRef CAS PubMed.
- J. S. Driscoll, G. F. Hazard, H. B. Wood and A. Goldin, Cancer Chemother. Rep., Part 2, 1974, 4, 1 CAS.
- A. J. Lin, R. S. Pardini, L. A. Cosby, B. J. Lillis, C. W. Shansky and A. C. Sartorelli, J. Med. Chem., 1973, 16, 1268 CrossRef CAS PubMed.
- E. Meggers, Curr. Opin. Chem. Biol., 2007, 11, 287 CrossRef CAS PubMed.
- C. G. Hartinger, S. Zorbas-Seifried, M. A. Jakupec, B. Kynast, H. Zorbas and B. K. Keppler, J. Inorg. Biochem., 2006, 100, 891 CrossRef CAS PubMed.
- J. M. Rademaker-Lakhai, D. Van Den Bongard, D. Pluim, J. H. Beijnen and M. Schellens, Clin. Cancer Res., 2004, 10, 3717 CrossRef CAS PubMed.
- M. A. Jakupec, V. B. Arion, S. Kapitza, E. Reisner, A. Eichinger, M. Pongratz, B. Marian, N. Graf, V. Keyserlingk and B. K. Keppler, Int. J. Clin. Pharmacol. Ther., 2005, 43, 595 CrossRef CAS PubMed.
- A. I. Vogel, Textbook of Practical Organic Chemistry, Longman, London, 5th edn, 1989 Search PubMed.
- N. Ahmad, J. J. Levison, S. D. Robinson and M. F. Uttley, Inorg. Synth., 1974, 15, 48 Search PubMed.
- R. A. S. Pelgado, W. Y. Lee, S. R. Choi, Y. Cho and M. J. Jun, Transition Met. Chem., 1991, 16, 241 CrossRef.
- J. Catalrin, F. Fabero, M. S. Guijarro, R. M. Claramunt, M. D. Santa Maria, M. de la Concepcidn Foces-Foces, F. Hernindez Cano, J. Elguero and R. Sastreil, J. Am. Chem. Soc., 1990, 112, 747 CrossRef.
- M. C. Altomare, M. Burla, G. Camalli, C. Cascarano, A. Giacovazzo, A. G. G. Guagliardi, G. Moliterni and R. Polidori, J. Appl. Crystallogr., 1999, 32, 115 CrossRef.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Crystallogr., 2008, 64, 112 CrossRef CAS PubMed.
- L. J. Farrugia, J. Appl. Crystallogr., 1997, 30, 565 CrossRef CAS.
- M. S. Blois, Nature, 1958, 29, 1199 CrossRef.
- T. Nash, J. Biochem., 1953, 55, 416 CrossRef CAS.
- L. C. Green, D. A. Wagner, J. Glogowski, P. L. Skipper, J. S. Wishnok and S. R. Tannenbaum, Anal. Biochem., 1982, 126, 131 CrossRef CAS PubMed.
- R. J. Ruch, S. J. Cheng and J. E. Klaunig, Carcinogenesis, 1989, 10, 1003 CrossRef CAS PubMed.
- C. Beauchamp and I. Fridovich, Anal. Biochem., 1971, 44, 276 CrossRef CAS PubMed.
- H. W. Lerner, G. Margraf, T. Kretz, O. Schiemann, J. W. Bats, G. Durner, F. Fabrizi de Biani, P. Zanello, M. Bolte and M. Z. Wagner, Z. Naturforsch., B: J. Chem. Sci., 2006, 61, 252 CAS.
- T. Sathiya Kamatchi, N. Chitrapriya, H. Lee, F. R. Fronczek and K. Natarajan, Dalton Trans., 2012, 41, 2066 RSC.
- F. Basuli, S. M. Peng and S. Bhattacharya, Inorg. Chem., 2001, 40, 1126 CrossRef CAS PubMed.
- A. K. Rappe, C. J. Casewit, K. S. Colwell, W. A. Goddard and W. M. Skiff, J. Am. Chem. Soc., 1992, 114, 10024 CrossRef CAS.
- M. D. Fryzuk, C. D. Montgomery and S. J. Rettig, Organometallics, 1991, 10, 467 CrossRef CAS.
- K. R. Flower and R. G. Pritchard, J. Organomet. Chem., 2001, 620, 60 CrossRef CAS.
- K. R. Flower, M. W. Garrould, L. G. Leal, C. Mangold, P. J. O. Malley and R. G. Pritchard, J. Organomet. Chem., 2008, 693, 408 CrossRef CAS.
- K. R. Flower and R. G. Pritchard, J. Organomet. Chem., 2001, 620, 60 CrossRef CAS.
- K. R. Flower, L. G. Leal and R. G. Pritchard, J. Organomet. Chem., 2005, 690, 3390 CrossRef CAS.
- K. N. Kumar, R. Ramesh and Y. Liu, J. Mol. Catal. A: Chem., 2007, 265, 218 CrossRef CAS.
- (a) E. C. Long and J. K. Barton, Acc. Chem. Res., 1990, 23, 271 CrossRef CAS; (b) R. F. Pasternack, E. J. Gibbs and J. J. Villafranca, Biochemistry, 1983, 22, 251 CrossRef.
- (a) F. Y. Wang, H. M. Chen, J. A. Parkinson, P. D. Murdoch and P. J. Sadler, Inorg. Chem., 2002, 41, 4509 CrossRef CAS PubMed; (b) F. Y. Wang, S. Weidt, J. J. Xu, C. L. Mackay, P. R. R. Langridge-Smith and P. J. Sadler, J. Am. Soc. Mass Spectrom., 2008, 19, 544 CrossRef CAS PubMed; (c) F. Y. Wang, J. J. Xu, A. Habtemariam, J. Bella and P. J. Sadler, J. Am. Chem. Soc., 2005, 127, 17734 CrossRef CAS PubMed; (d) S. Maity, S. Hattacharya and S. Chaudhury, Chemosphere, 2009, 77, 319 CrossRef CAS PubMed.
- H. Y. Liu, Z. H. Xu, X. H. Liu, P. X. Xi and Z. Z. Zeng, Chem. Pharm. Bull., 2009, 57, 1237 CrossRef CAS PubMed; Y. Hu, Y. Yang, C. Dai, Y. Liu and X. Xiao, Biomacromolecules, 2010, 11, 106 CrossRef PubMed.
- A. Sulkowska, J. Mol. Struct., 2002, 614, 227 CrossRef CAS.
- Y. Wang, H. Zhang, G. Zhang, W. Tao and S. Tang, J. Lumin., 2007, 126, 211 CrossRef CAS.
- G. Z. Chen, X. Z. Huang, J. G. Xu, Z. B. Wang and Z. Z. Zhang, Method of Fluorescent Analysis, Science Press, Beijing, 2nd edn, 1990, p. 123, 126, ch, 4 Search PubMed.
- J. N. Miller, Proc. Anal. Div. Chem. Soc., 1979, 16, 203 CAS.
- J. H. Tang, F. Luan and X. G. Chen, Bioorg. Med. Chem., 2006, 49, 3210 CrossRef PubMed.
- B. Liu, Y. Guo, J. Wang, R. Xu, R. Wang, L. Q. Zhang and Y. Xu, J. Lumin., 2010, 130, 1036 CrossRef CAS.
- K. Tsai, T. G. Hsu, K. M. Hsu, H. Cheng, T. Y. Liu, C. F. Hsu and C. W. Kong, Free Radical Biol. Med., 2001, 31, 1465–1472 CrossRef CAS PubMed.
- C. S. Rivas, J. C. Espin and H. Wichers, Phytochem. Anal., 2000, 11, 330 CrossRef.
- (a) H. L. Huang, Y. L. Liu, C. H. Zeng, J. H. Yao, Z. H. Liang, Z. Z. Li and F. H. Wu, J. Mol. Struct., 2010, 966, 136 CrossRef CAS; (b) Y. L. Liu, C. H. Zeng, Z. H. Liang, J. H. Yao, H. L. Huang, Z. Z. Li and F. H. Wu, Eur. J. Med. Chem., 2010, 45, 3087 CrossRef CAS PubMed; (c) X. L. Hong, H. Li and C. H. Peng, J. Mol. Struct., 2011, 990, 197 CrossRef CAS; (d) Z. Z. Li, Z. H. Liang, H. L. Huang and Y. J. Liu, J. Mol. Struct., 2011, 1001, 36 CrossRef CAS; (e) Y. J. Liu, Z. H. Liang, Z. Z. Li, J. H. Yao and H. L. Huang, J. Organomet. Chem., 2011, 696, 2728 CrossRef CAS.
- T. Sathiya Kamatchi, N. Chitrapriya, H. Lee, F. R. Fronczek and K. Natarajan, Eur. J. Med. Chem., 2013, 59, 253 CrossRef CAS PubMed.
- X. Yanga, L. Chenb, Y. Liua, Y. Yanga, T. Chena, W. Zhenga, J. Liua and Q. Y. He, Biochimie, 2012, 94, 345 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: 1H-NMR spectrum of ligand LH, 2,5-di(1H-pyrazol-1-yl)-1,4-benzoquinone (Fig. S1); 1H-NMR spectrum of the complex 3 (Fig. S2); 1H-NMR spectrum of the complex 4 (Fig. S3); 13C-NMR spectrum of the complex 3 (Fig. S4); 13C-NMR spectrum of the complex 4 (Fig. S5); packing diagram of the unit cell for complex 3 (Fig. S6); packing diagram of the unit cell for complex 4 (Fig. S7); the emission spectra of the DNA–EB system (λexc = 515 nm, λem = 530–750 nm), in the presence of the ligand LH and complexes 1–4 (Fig. S8); synchronous spectra of BSA (1 μM) in the presence of increasing amounts of the ligand LH and complexes 1–4 for a wavelength difference of Δλ = 15 nm (Fig. S9). CCDC 797114 and 797115. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c6ra05867a |
| This journal is © The Royal Society of Chemistry 2016 |