 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Targeting of the Leishmania mexicana cysteine protease CPB2.8ΔCTE by decorated fused benzo[b]thiophene scaffold†
A.
Scala
a,
N.
Micale
a,
A.
Piperno
a,
A.
Rescifina
 b,
T.
Schirmeister
c,
J.
Kesselring
c and
G.
Grassi
*a
b,
T.
Schirmeister
c,
J.
Kesselring
c and
G.
Grassi
*a
aDipartimento di Scienze Chimiche, Biologiche, Farmaceutiche ed Ambientali, Università di Messina, V.le F. Stagno D'Alcontres 31, 98166 Messina, Italy. E-mail: ggrassi@unime.it
bDipartimento di Scienze del Farmaco, Università degli Studi di Catania, V.le A. Doria, 95125 Catania, Italy
cInstitute of Pharmacy and Biochemistry, University of Mainz, Staudinger Weg 5, D 55099 Mainz, Germany
First published on 11th March 2016
Abstract
A potent and highly selective anhydride-based inhibitor of Leishmania mexicana cysteine protease CPB2.8ΔCTE (IC50 = 3.7 μM) was identified. The details of the interaction of the ligand with the enzyme active site were investigated by NMR biomimetic experiments and docking studies. Results of inhibition assays, NMR and theoretical studies indicate that the ligand acts initially as a non-covalent inhibitor and later as an irreversible covalent inhibitor by chemoselective attack of CYS 25 thiolate to an anhydride carbonyl.
Introduction
Despite progress made in both the basic knowledge of many infectious diseases and drug discovery and development, tropical infectious diseases such as leishmaniasis, malaria, trypanosomiasis, and Chagas' disease, continue to cause significant morbidity and mortality predominantly in the less developed world.1Leishmaniasis is one of the major tropical diseases, ranking second only to malaria for mortality rate,1 caused by protozoan parasites of the genus Leishmania.2 Depending on the tropism, the disease is characterized by three different clinical forms: visceral, cutaneous, and mucocutaneous. The visceral form is fatal in 85–90% of untreated cases. No effective vaccine is available against leishmaniasis, therefore chemotherapy is the only effective way to treat all forms of this neglected disease.3 Current therapy relies mainly on drugs that were developed decades ago, such as pentavalent antimony agents, amphotericin B, paromomycin and pentamidine. Severe toxic effects combined with the emergence of drug-resistant parasite strains has created an urgent and continuous need for new, safe and efficacious drugs.
Cysteine proteases (CPs) play crucial roles in the biology of parasites and their inhibition is emerging as an important strategy to combat parasitic diseases.4,5 Leishmania (L.) protozoa express high levels of several classes of CPs belonging to the papain family, that are crucial to parasite metabolism, reproduction and intracellular survival.6 In particular L. mexicana possesses three CPs of the papain superfamily, namely CPA and CPB, both of which are cathepsin L-like, and CPC, which is cathepsin B-like.7 Inhibitors of the CPBs isoenzymes have been shown to reduce the infectivity of L. mexicana both in vitro6 and in vivo,7 thus providing further evidences that these CPBs isoenzymes are virulence factors. To the best of our knowledge, only a few reports describe the identification of novel CPBs inhibitors,8–10 as the CPBs are relatively unexplored drug targets. They can be divided in three broad groups: natural compounds (e.g. morelloflavones), metal complexes (such as tellurium, palladium, and gold derivatives) and CPBs inhibitors endowed with an electrophilic warhead.11,12 The latter group can be further subdivided in peptidic and non-peptidic CPBs inhibitors, which are labelled as α-ketoheterocycles 1,10 thiosemicarbazones 2,8 semicarbazones 38 and nitriles 48 (Fig. 1), according to their warhead-types that interacts with the cysteine thiolate of the active site.13,14
 | ||
| Fig. 1 Structures of CPB2.8 inhibitors: α-ketoheterocycles 1, thiosemicarbazones 2, semicarbazones 3, nitriles 4 and the anhydride-based inhibitor 5 reported herein. | ||
As a part of an ongoing program of targeting small molecular weight heterocyclic scaffolds, the activity of a fused benzo[b]thiophene derivative 5 (Fig. 1), whose synthesis15 and properties as ionophore16 we have already reported, was tested against a panel of human and parasitic CPs, including the mature recombinant form of the amastigote-specific isoform CPB2.8 of L. mexicana cysteine protease, expressed without the C-terminal extension and so designed mature CPB2.8ΔCTE, and the rhodesain and human cathepsin-B and -L.
The choose of biological targets was done on the basis of a combined PharmMapper17 and wwLigCSRre 3D ligand-based18 servers approach, followed by a polish up by Tanimoto similarity index search.19 This strategy allowed us to identify the bicyclic fused dihydropyrrolo[3,2-c]isoxazol-6-one core, belonging to a series of compounds acting as cysteinyl proteinase inhibitors,20 as the best match to the tetrahydrofuro[3,4-b]pyrrole-4,6-dione core of compound 5.
From this biological screening compound 5 turned out to be active against mature L. mexicana cysteine protease CPB2.8ΔCTE, with a high selectivity for the parasite's enzyme with respect to the highly similar human CPs.
Results and discussion
The synthesis15 of the target compound 5 (Scheme 1) relies on an intriguing 1,3-dipolar cycloaddition between thiocoumarin 6 having an electron-withdrawing group at the C-3 position and 3,4-dimethyl-2-phenyl-1,3-oxazolium-5-olate 7, a powerful mesoionic compound, toward which our interest for the synthesis of highly substituted heterocycles is well known.21–23The preliminary screening at 20 μM of 5 against mature CPB2.8ΔCTE produced a remarkable inhibition (≈90%) of the target enzyme (Table 1). No inhibition (n.i.) was detected in the screening against the other parasitic CPs at our disposal (i.e. rhodesain, Table 1) and, more importantly, no significant cross-reactivity was detected towards highly similar human CPs (Table 1) such as cathepsin-B (n.i. at 20 μM) and cathepsin-L (only ≈30% of inhibition at 20 μM) suggesting that 5 selectively interacts with the target, realistically due to its highly conformationally constrained structure.
Therefore, compound 5 was further evaluated by progress curve analysis24 (Fig. 2 and 3) using a continuous readout.
Compound 5 has a non-peptidic tetracyclic scaffold with two electrophilic moieties, a peripheral cyclic anhydride and a lateral ketone group, both warhead suitable for nucleophilic attack by the CYS 25 thiolate of the enzyme.
Inhibition of CPs can take place via several different methods, including covalent inhibition, blockage or distortion of the catalytic active site via competition with non-covalent inhibitors.
Progress curves for the inhibition of mature CPB2.8ΔCTE by 5 measured over a time period of 10 min (Fig. 2) indicated a non-time-dependent mechanism, suggesting a non-covalent or fast-binding covalent reversible inhibition mechanism.
However, the chemical structure of our hit compound with the presence of two reactive warheads that could undergo covalent modification by the active site thiol of the enzyme suggest an irreversible binding mechanism.
In contrast to these 10 min assays the measurement of enzyme activity over a time period of 30 min showed time-dependent inhibition. Thus, these two experiments did not unequivocally prove the inhibition mechanism. In order to clarify the mode of action, dialysis assays were performed (see below).
The assays with 10 min measurement time yielded an IC50 of 3.7 μM. With the substrate concentration used (10 μM) and the Km value of 5.0 μM, the Ki value has been calculated to 1.2 μM (according to Cheng–Prusoff equation for competitive inhibitors in a classic mode).25,26
With the progress curves obtained over a 30 min time period (Fig. 3) first-order rate constants of inhibition (kobs values) were obtained which were fitted to the inhibitor concentrations24 yielding a second-order rate constant of inhibition, k2nd, of 4290 ± 5 M−1 s−1 with a Ki value of 1.36 ± 0.075 μM.
To clarify the issue of irreversibility of inhibition, dialysis assays by mixing enzyme and inhibitor 5 were performed in order to check if the enzyme activity could be recovered after this treatment. As shown in Fig. 4, the dialysis did not lead to the regeneration of enzyme activity, proving the irreversible inhibition which fits with the order of reactivity of the electrophilic moieties of our compound. All together these outcomes suggested that 5 inhibits the target by two inhibition pathways, a reversible fast one and an irreversible one.
Selectivity and potency will combine to make 5 eligible as a new lead structure for the development of anti-leishmanial agents. In addition, 5 might offer advantages in term of metabolic stability comparing to peptide inhibitors reported in literature, whose structures typically rely on a cleavable (pseudo) peptide recognition motif bearing a C-terminal electrophilic warhead.27–29
NMR biomimetic experiments (Fig. 5) were performed in DMSOd6/D2O 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 using N-(tert-butoxycarbonyl)-cysteine methyl ester to shed light on the interaction modes of 5 with the target cysteine protease. Therefore, N-(tert-butoxycarbonyl)-cysteine methyl ester was added to DMSOd6/D2O solution of 5 in a nearly equimolar ratio. The reaction was monitored at different times directly in the NMR tube at a probe temperature of 25 °C, observing the shift and the appearance/disappearance of selected signals.
2 using N-(tert-butoxycarbonyl)-cysteine methyl ester to shed light on the interaction modes of 5 with the target cysteine protease. Therefore, N-(tert-butoxycarbonyl)-cysteine methyl ester was added to DMSOd6/D2O solution of 5 in a nearly equimolar ratio. The reaction was monitored at different times directly in the NMR tube at a probe temperature of 25 °C, observing the shift and the appearance/disappearance of selected signals.
NMR studies reveal that 5 chemoselectively reacts with the cysteine sulfhydryl group by ring-opening of the anhydride moiety (Fig. 5A). In particular the 13C NMR spectrum (Fig. 5B) shows the disappearance of the resonance a at 167.4 ppm (attributed to the carbon of the anhydride next to the ketone group by gHMBCAD experiments), a slight shift of the carbonyl signal b from 171.3 to 171.8 (b′), and the appearance of two new signals at 181.6 ppm and 171.7 ppm, related to the newly formed thioester carbonyl group (a′) and to the CO2Me of the cysteine methyl ester, respectively. A slight downfield shift of the signal of ketone carbonyl was observed from 192.6 to 193.7 ppm.
These findings support the irreversible conversion of ligand 5 into compound 8. 1H NMR spectra show the presence of a complex signal pattern, due to the presence of more than one product, according to the multiplicity of the reactive sites. Nevertheless, an unambiguous set of signals are consistent with the formation of 8 as the main product. In particular, arrayed experiments (Fig. 5C) clearly revealed the upfield shift of the signal related to the hydrogen of the thienopyrrole from 4.9 (c) to 4.0 ppm (c′), and the downfield shift of both the cysteine CH from 4.1 (d) to 4.2 (d′) ppm and of the cysteine CH2 from 2.6–2.8 (e) to 2.8–3.1 (e′) ppm. The structure of 8 was unambiguously assigned on the basis of COSY, gHSQCAD and gHMBCAD experiments.
To further rationalize the experimental findings, homology modeling, molecular dynamics (MD), and noncovalent and covalent docking experiments were carried out. In order to get the 3D structure of the protein for MD and docking studies, a structural model of active mature L. mexicana CPB2.8ΔCTE was generated with the aid of YASARA Structure software.30 Starting from the target sequence possible templates were identified by running three PSIBLAST iterations to extract a position specific scoring matrix from UniRef90, and then searching the PDB for a match (i.e. hits with an E-value below the homology modeling cutoff of 0.5). The best result was the crystal structure of cruzain bound to vinyl sulfone analog of Wrr-483 (PDB ID 4PI3) with a resolution of 1.27 Å; this template was downloaded from PDB_REDO database,31 since re-refinement improved the structure quality Z-score by 0.090. The sequence identity of 60.3% and the sequence similarity of 74.8% between mature CPB2.8ΔCTE and cruzain was reasonable for the generation of a qualified homology model. Then a full unrestrained simulated annealing minimization was run for the entire model and this fully refined model has been accepted as the final one (see ESI† for model validation) and used as starting point for successive studies. The so obtained homology model showed a Cα RMSD value of 0.42 Å compared to its template structure. The inactivation of a protease by an active-site directed irreversible inhibitor usually proceeds by the rapid formation of a non-covalent reversible enzyme–inhibitor complex (E–I). Successively, in a slower chemical step, a covalent bond is formed with the enzyme to generate the enzyme–inhibitor adduct (E–I).32
So, we conducted the study utilizing this sequence: (i) non-covalent docking of ligand upon mature CPB2.8DCTE enzyme; (ii) 40 ns of MD simulation of the best pose obtained for ligand–CPB2.8ΔCTE complex, to accommodate the ligand; (iii) non-covalent re-docking of the complex obtained from the last 3 ns of MD simulation averaged frames; (iv) 40 ns of MD simulation of covalent docked ligand, based on the best re-docked pose; (v) covalent docking of the complex obtained from the last 3 ns of MD simulation averaged frames, to assess the best conformation for the free-moiety of the ligand; (vi) 400 ns of MD simulation of the best non-covalent docked complex, to verify the stability and the correctness of the complex (Fig. S1–3†). To validate the homology model in performing a suitable level of docking accuracy we successfully docked two well-known CPB2.8ΔCTE inhibitors with different Ki values (Fig. S4 and Table S1†).
Since compound 5 exists as a racemic mixture and it is a tertiary amine, considering physiological conditions (pH = 7.2), we performed the first phase on both N-protonated enantiomers, using the most stable protonated diastereomer.
The enantiomer 5-H, with configuration 3aS,4R,4aR,9bR,9cR (Fig. 6A), resulted the best ligand, with a difference in the free energy of binding (ΔGB) of 0.6 kcal mol−1, and with a pose suitable for successive covalent docking, contrarily to its enantiomer. Then, all further studies were conducted on (3aS,4R,4aR,9bR,9cR)-5-H enantiomer, simply mentioned as 5-H. Moreover, to computationally determine the best electrophilic site of 5-H for the nucleophilic attach of cysteine, considering the possibility that this reaction could be either under orbital or charge control, we fully optimized compound 5-H in water, at DFT level of theory, to retrieve the shape and localization of the lowest unoccupied molecular orbital (LUMO) and the charge distribution under natural bond orbital (NBO) scheme. As depicted in Fig. 6B the LUMO is centered on the Ca carboxylic moiety and the highest positive charge is located on the same atom; this is perfectly in accord to the result obtained by NMR biomimetic experiments.
 | ||
| Fig. 6 (A) Structure of compound (3aS,4R,4aR,9bR,9cR)-5-H with numeration. (B) 3D molecular model of 5-H with LUMO and selected NBO charges. | ||
The results of the non-covalent re-docking of 5-H showed that it bounds the enzyme exposing both carbonyl and carboxyl moieties to the nucleophilic cysteine 25 residue, that is, as well-known,32 involved in the instauration of the covalent bond (Fig. 7A–C). As regards to the water environment no one molecule is directly involved in ligand–receptor complex stabilization; however, a water molecule establish a hydrogen bond with the ammonium of ligand (Fig. 7B). The calculated ΔGb of −7.3 kcal mol−1 corresponds to a Ki of 4.4 μM that is, in itself, well in agreement with the experimental one. Hydrogen bond interactions and their energies, according to Fig. 7A, have been reported in ESI (Table S2†). The covalent docking performed starting from the best non-covalent docked pose showed that compound 5-H results even more deeply embedded in the active site with both carboxylic moieties engaged in hydrogen bonds with GLN 19, CYS 25, and TRP 185 residues (Fig. 7D). These results parallel the experimental ones: the ligand acts immediately as non-covalent inhibitor, and then the system is engaged in the formation of the covalent bond. Moreover, it is possible to note both in non-covalent and covalent pose of docked ligand that there is an empty groove in the protein (Fig. 7C and D), that can be further exploited to construct a more performant ligand.
 | ||
| Fig. 7 (A) 2D sketch interactions of non-covalent docked pose of 5-H. (B) 3D representation as in A, including surrounding water molecules. (C) 3D mapped surface of CPB2.8ΔCTE with 5-H non-covalent docked; CYS 25 overlooks on both carbonyl and carboxyl moieties. (D) Covalent docked pose of 5-H with the CYS 25 bounded to the carboxyl moiety a (according to the numbering in Fig. 6A) of the anhydride. | ||
Finally, to deeply investigate the interactions involved in the recognition process we performed a computational study using the quantum mechanics/molecular mechanics (QM/MM) approach, the so called ONIOM method,33 on the wholly solvated ligand–enzyme complex, including the residues surrounding the ligand in the quantum mechanical layer of the calculation. In this case, the LUMO of the entire system is centered only on ligand but, compared to the result obtained on the isolated ligand, now it is extended to the Cb carboxylic moiety (Fig. 8). However, it is evident from Fig. 8 that the HOMO −10, centered on sulfur atom of CYS 25, is in the correct position to the subsequent nucleophilic attach to the Ca. Interestingly, the complete optimization of the entire ONIOM system brings to the formation of the intermediate of the classical two step acyl substitution reaction and it is a computational evidence that the system spontaneously evolves towards the covalent interaction (see video in ESI†).
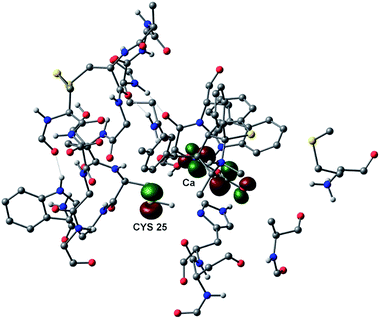 | ||
| Fig. 8 ONIOM 3D molecular model of 5-H–CPB2.8ΔCTE complex with HOMO −10 and LUMO. It is showed only the QM layer; non-polar hydrogens are omitted for clarity. | ||
Experimental
Enzyme assays
The preliminary screening against CPs was performed with 20 μM inhibitor concentrations by using an equivalent amount of DMSO as a negative control. Recombinant enzymes (i.e. rhodesain and mature L. mexicana CPB2.8ΔCTE) were expressed as previously described.34,35 Product release from substrate hydrolysis (Cbz-Phe-Arg-AMC, 10 μM or 20 μM) was determined continuously over a period of 10 min or 30 min and the fluorescence measured using an Infinite 200 PRO microplate reader (Tecan, Männedorf, Switzerland) at 30 °C with a 380 nm excitation filter and a 460 nm emission filter. The higher substrate concentration of 20 μM was chosen to ensure linear increase of the fluorescent product AMC over time in absence of the inhibitor during 30 min. Rhodesain was incubated at 20 °C with test compound in a 50 mM sodium acetate buffer (pH 5.5) containing 10 mM 1,4-dithiothreitol (DTT), 5 mM EDTA, and 200 mM NaCl, whereas L. mexicana CPB2.8ΔCTE was incubated in the same conditions by using a 50 mM sodium acetate buffer (pH 6.5) containing 5 mM DTT and 5 mM EDTA. Cathepsins-B and -L were purchased from Calbiochem and the assays were performed as previously described.36 Also in this case Cbz-Phe-Arg-AMC was used as the substrate (80 μM for cathepsin-B; 5 μM for cathepsin-L).Dialysis assays
Mature L. mexicana CPB2.8ΔCTE (13 μg mL−1) was incubated with 5 (50 μM) for 5 min. The reaction mixture was then subjected to dialysis against reaction buffer (5000-fold excess) for 3 h. The residual enzyme activities were determined by adding substrate (10 μM). The enzyme was subjected to the same procedure in the absence of inhibitor in order to confirm its stability to the dialysis conditions. Positive and negative control assays, i.e. dialysis with fully irreversible (E-64) and fully reversible inhibitors,37 were also performed.NMR biomimetic experiments
NMR measurements were performed on a Varian 500 (500 MHz for 1H; 125 MHz for 13C) spectrometer at 25 °C using DMSOd6/D2O 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 as solvent (D2O was phosphate buffered at pH 7.2). Chemical shifts are expressed in ppm respect to TMS. The NMR sample was prepared by dissolving compound 5 (4 mg, 0.008 mmol) in 0.6 mL of DMSOd6/D2O 8
2 as solvent (D2O was phosphate buffered at pH 7.2). Chemical shifts are expressed in ppm respect to TMS. The NMR sample was prepared by dissolving compound 5 (4 mg, 0.008 mmol) in 0.6 mL of DMSOd6/D2O 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, then 2 mg of N-(tert-butoxycarbonyl)-cysteine methyl ester (0.008 mmol) were added. The biomimetic experiments were carried out directly in the NMR tube by recording 1H NMR spectra every 30 min for 48 h. Afterwards 13C NMR spectrum was registered, and finally gCOSY, gHSQCAD, and gHMBCAD experiments were run for the determination of the structure of compound 8.
2, then 2 mg of N-(tert-butoxycarbonyl)-cysteine methyl ester (0.008 mmol) were added. The biomimetic experiments were carried out directly in the NMR tube by recording 1H NMR spectra every 30 min for 48 h. Afterwards 13C NMR spectrum was registered, and finally gCOSY, gHSQCAD, and gHMBCAD experiments were run for the determination of the structure of compound 8.
Preparation of ligands, DFT and ONIOM calculations
The 3D structures of ligands were built using Winmostar (5.009) software38 and all geometries were fully optimized, in the same software, with the semi empirical PM6 (ref. 39) Hamiltonian implemented in MOPAC2012 (15.156 W).40 The optimized structure of 5-H was further fully optimized, in water, into the DFT framework, at the M06-2X/cc-pvtz level of theory, utilizing the implicit polarizable continuum model for solvation. The electronic structure of 5-H was studied by the NBO method. All the optimizations were carried out by Berny's analytic gradient method as implemented in Gaussian 09 suite.41The YASARA2 optimized geometry of the best re-docked pose obtained by point iv, in the above described sequence, was used as a starting point for the QM/MM calculations using a two-layer ONIOM method.33 The TAO package42 was used to prepare the system and the parmchk module of antechamber package43 to retrieve non-standard force field parameters. The ligand and 17 surrounding amino acid fragments (GLN 19, GLY 20, CYS 22, GLY 23, SER 24, CYS 25, CYS 63, ASP 64, GLY 65, GLY 66, ALA 142, PHE 145, MET 146, ASN 162, HIS 163, GLY 164 and TRP 185) were included in the QM region (349 atoms). The QM part of the system was described at the M06-2X/6-31G(d,p) level of density functional theory for optimization and at M06-2X/cc-pvtz one for single point calculations. The MM part of the system was described using the parm96 parameters of the AMBER force field,44 as implemented in Gaussian 09, and includes the remaining amino acids and 6338 water molecules (only water molecules extending 3 Å from the surface of the complex were retained).
Molecular dynamics simulations
The molecular dynamics simulations of the mature CPB2.8ΔCTE/ligand complexes (based on the PDBs prepared as described above) were performed with the YASARA Structure package (15.3.8).30 A periodic simulation cell with boundaries extending 10 Å from the surface of the complex was employed. The box was filled with water, with a maximum sum of all bumps per water of 1.0 Å, and a density of 0.997 g mL−1 with explicit solvent. YASARA's pKa utility was used to assign pKa values at pH 7.2,45 and the cell was neutralized with NaCl (0.9% by mass); in these conditions 5-H ligand results protonated at pyrrolidinic N-Me. Waters were deleted to readjust the solvent density to 0.997 g mL−1. The YASARA2 force field was used with long-range electrostatic potentials calculated with the Particle Mesh Ewald (PME) method, with a cutoff of 8.0 Å.44,46,47 The ligand force field parameters were generated with the AutoSMILES utility,48 which employs semiempirical AM1 geometry optimization and assignment of charges, followed by assignment of the AM1BCC atom and bond types with refinement using the RESP charges, and finally the assignments of general AMBER force field atom types. Optimization of the hydrogen bond network of the various enzyme–ligand complexes was obtained using the method established by Hooft et al.,49 in order to address ambiguities arising from multiple side chain conformations and protonation states that are not well resolved in the electron density.50 A short MD was run on the solvent only. The entire system was then energy minimized using first a steepest descent minimization to remove conformational stress, followed by a simulated annealing minimization until convergence (<0.01 kcal mol−1 Å−1). The MD simulation was then initiated, using the NVT ensemble at 298 K, and integration time steps for intramolecular and intermolecular forces every 1.25 fs and 2.5 fs, respectively. The MD simulation was stopped after 40 ns and, on the averaged structure of the last 3 ns frames, a second cycle of energy minimization, identical to the first, was applied.Docking protocol
Macromolecules and ligands, as obtained after MD simulation and energy minimization, were prepared with Vega ZZ51 (3.0.3.18) assigning Gasteiger charges to protein and AM1BCC ones to ligand. Docking was performed with AutoDock Vina (1.1.2) software.52 Because no water molecule is directly involved in complex stabilization they were not considered in the docking process. All protein amino acidic residues were kept rigid whereas all single bonds of ligands were treated as full flexible. The ligand box was centered at x = 9.32, y = −5.95 and z = −7.50 coordinates with a grid size of 15 × 20 × 18 Å and the exhaustiveness parameter related to Lamarckian genetic algorithm was set to 15.Conclusions
In summary, we have discovered a potent and highly selective anhydride-based inhibitor of mature L. mexicana cysteine protease CPB2.8ΔCTE, with an Ki of ca. 1.3 μM, that possess no significant cross-reactivity towards highly similar human CPs such as cathepsin-B and cathepsin-L. The details of the catalytic reaction mechanism of ligand with the enzyme active site amino acids were investigated by NMR biomimetic experiments and docking studies. All results indicate that the ligand acts immediately as non-covalent inhibitor and then as irreversible covalent inhibitor by chemoselective attack of CYS 25 thiolate to Ca anhydride carbonyl. Moreover, docking studies suggest that compound (3aS,4R,4aR,9bR,9cR)-5-H can be considered the eutomer with a modest eudysmic ratio of 3. Finally, the inspection of 5-H–CPB2.8ΔCTE complex molecular surface account for a fine tuning of ligand substituents to further improve both potency and selectivity.Acknowledgements
This work was partially supported by MIUR [project PRIN 20109Z2XRJ_010]; we thank Jeremy C. Mottram for kindly providing the LmCPB2.8 protease.Notes and references
- World Health Organization (WHO), World Health Report, 2002 Search PubMed.
- M. den Boer, D. Argaw, J. Jannin and J. Alvar, Clin. Microbiol. Infect., 2011, 17, 1471–1477 CrossRef CAS PubMed.
- J. N. Sangshetti, F. A. K. Khan, A. A. Kulkarni, R. Arote and R. H. Patil, RSC Adv., 2015, 5, 32376–32415 RSC.
- C. R. Caffrey, A. P. Lima and D. Steverding, Adv. Exp. Med. Biol., 2011, 712, 84–99 CrossRef CAS PubMed.
- N. Kopitar-Jerala, Curr. Protein Pept. Sci., 2012, 13, 767–775 CrossRef CAS PubMed.
- M. J. Frame, J. C. Mottram and G. H. Coombs, Parasitology, 2000, 121, 367–377 CrossRef CAS PubMed.
- M. A. Juliano, D. R. Brooks, P. M. Selzer, H. L. Pandolfo, W. A. S. Judice, L. Juliano, M. Meldal, S. J. Sanderson, J. C. Mottram and G. H. Coombs, Eur. J. Biochem., 2004, 271, 3704–3714 CrossRef CAS PubMed.
- J. Schröder, S. Noack, R. J. Marhöfer, J. C. Mottram, G. H. Coombs and P. M. Selzer, PLoS One, 2013, 8, e77460 Search PubMed.
- P. M. Selzer, S. Pingel, I. Hsieh, B. Ugele, V. J. Chan, J. C. Engel, M. Bogyo, D. G. Russell, J. A. Sakanari and J. H. McKerrow, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 11015–11022 CrossRef CAS.
- K. Steert, M. Berg, J. C. Mottram, G. D. Westrop, G. H. Coombs, P. Cos, L. Maes, J. Joossens, P. Van der Veken, A. Haemers and K. Augustyns, ChemMedChem, 2010, 5, 1734–1748 CrossRef CAS PubMed.
- A. S. Nagle, S. Khare, A. B. Kumar, F. Supek, A. Buchynskyy, C. J. N. Mathison, N. K. Chennamaneni, N. Pendem, F. S. Buckner, M. H. Gelb and V. Molteni, Chem. Rev., 2014, 114, 11305–11347 CrossRef CAS PubMed.
- M. Navarro, C. Gabbiani, L. Messori and D. Gambino, Drug Discovery Today, 2010, 15, 1070–1078 CrossRef CAS PubMed.
- I. J. Gonzalez-Leal, B. Roger, A. Schwarz, T. Schirmeister, T. Reinheckel, M. B. Lutz and H. Moll, PLoS Neglected Trop. Dis., 2014, 8, e3194 Search PubMed.
- U. Schurigt, C. Schad, C. Glowa, U. Baum, K. Thomale, J. K. Schnitzer, M. Schultheis, N. Schaschke, T. Schirmeister and H. Moll, Antimicrob. Agents Chemother., 2010, 54, 5028–5041 CrossRef CAS PubMed.
- M. Cordaro, G. Grassi, A. Rescifina, U. Chiacchio, F. Risitano and A. Scala, Tetrahedron, 2011, 67, 608–611 CrossRef CAS.
- M. Cordaro, G. Grassi, A. Rescifina, U. Chiacchio, F. Risitano and A. Scala, J. Mol. Struct., 2011, 991, 143–148 CrossRef CAS.
- X. F. Liu, S. S. Ouyang, B. A. Yu, Y. B. Liu, K. Huang, J. Y. Gong, S. Y. Zheng, Z. H. Li, H. L. Li and H. L. Jiang, Nucleic Acids Res., 2010, 38, W609–W614 CrossRef CAS PubMed.
- O. Sperandio, M. Petitjean and P. Tuffery, Nucleic Acids Res., 2009, 37, W504–W509 CrossRef CAS PubMed.
- D. Bajusz, A. Racz and K. Heberger, J. Cheminf., 2015, 7, 1–13 CAS.
- Y. K. Wang, A. Benn, N. Flinn, T. Monk, M. Ramjee, J. Watts and M. Quibell, Bioorg. Med. Chem. Lett., 2005, 15, 1327–1331 CrossRef CAS PubMed.
- M. Cordaro, G. Grassi, F. Risitano and A. Scala, Synlett, 2009, 103–105, DOI:10.1055/s-0028-1087483.
- M. Cordaro, G. Grassi, F. Risitano and A. Scala, Tetrahedron, 2010, 66, 2713–2717 CrossRef CAS.
- A. Piperno, A. Scala, F. Risitano and G. Grassi, Curr. Org. Chem., 2014, 18, 2691–2710 CrossRef CAS.
- W. X. Tian and C. L. Tsou, Biochemistry, 1982, 21, 1028–1032 CrossRef CAS PubMed.
- R. Z. Cer, U. Mudunuri, R. Stephens and F. J. Lebeda, Nucleic Acids Res., 2009, 37, W441–W445 CrossRef CAS PubMed.
- C. Yung-Chi and W. H. Prusoff, Biochem. Pharmacol., 1973, 22, 3099–3108 CrossRef.
- R. Ettari, F. Bova, M. Zappalà, S. Grasso and N. Micale, Med. Res. Rev., 2010, 30, 136–167 CAS.
- R. Ettari, L. Tamborini, I. C. Angelo, N. Micale, A. Pinto, C. De Micheli and P. Conti, J. Med. Chem., 2013, 56, 5637–5658 CrossRef CAS PubMed.
- M. Frizler, M. Stirnberg, M. T. Sisay and M. Gutschow, Curr. Top. Med. Chem., 2010, 10, 294–322 CrossRef CAS PubMed.
- E. Krieger, YASARA, 13.9.8, ed. YASARA Biosciences GmbH, Vienna, Austria, 2013 Search PubMed.
- R. P. Joosten, K. Joosten, G. N. Murshudov and A. Perrakis, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2012, 68, 484–496 CrossRef CAS PubMed.
- J. C. Powers, J. L. Asgian, O. D. Ekici and K. E. James, Chem. Rev., 2002, 102, 4639–4750 CrossRef CAS PubMed.
- F. Clemente, T. Vreven, and M. J. Frisch, in Quantum Biochemistry, ed. C. Matta, Wiley VCH, Weinheim, 2010, pp. 61–84 Search PubMed.
- C. R. Caffrey, E. Hansell, K. D. Lucas, L. S. Brinen, A. A. Hernandez, J. N. Cheng, S. L. Gwaltney, W. R. Roush, Y. D. Stierhof, M. Bogyo, D. Steverding and J. H. McKerrow, Mol. Biochem. Parasitol., 2001, 118, 61–73 CrossRef CAS PubMed.
- S. J. Sanderson, K. G. J. Pollock, J. D. Hilley, M. Meldal, P. St Hilaire, M. A. Juliano, L. Juliano, J. C. Mottram and G. H. Coombs, Biochem. J., 2000, 347, 383–388 CrossRef CAS PubMed.
- R. Vicik, M. Busemann, C. Gelhaus, N. Stiefl, J. Scheiber, W. Schmitz, F. Schulz, M. Mladenovic, B. Engels, M. Leippe, K. Baumann and T. Schirmeister, ChemMedChem, 2006, 1, 1126–1141 CrossRef CAS PubMed.
- L. R. de Sousa, H. Wu, L. Nebo, J. B. Fernandes, M. F. da Silva, W. Kiefer, T. Schirmeister and P. C. Vieira, Exp. Parasitol., 2015, 156, 42–48 CrossRef CAS PubMed.
- N. Senda, Idemitsu Gihou, 2006, 49, 106–111 Search PubMed.
- J. J. P. Stewart, J. Mol. Model., 2007, 13, 1173–1213 CrossRef CAS PubMed.
- J. J. P. Stewart, MOPAC2012, http://OpenMOPAC.net Search PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery, J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, Ö. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09 Rev. D.01 Search PubMed.
- P. Tao and H. B. Schlegel, J. Comput. Chem., 2010, 31, 2363–2369 CAS.
- J. M. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2005, 26, 114 CrossRef CAS.
- W. D. Cornell, P. Cieplak, C. I. Bayly, I. R. Gould, K. M. Merz, D. M. Ferguson, D. C. Spellmeyer, T. Fox, J. W. Caldwell and P. A. Kollman, J. Am. Chem. Soc., 1995, 117, 5179–5197 CrossRef CAS.
- E. Krieger, J. E. Nielsen, C. A. E. M. Spronk and G. Vriend, J. Mol. Graphics Modell., 2006, 25, 481–486 CrossRef CAS PubMed.
- Y. Duan, C. Wu, S. Chowdhury, M. C. Lee, G. M. Xiong, W. Zhang, R. Yang, P. Cieplak, R. Luo, T. Lee, J. Caldwell, J. M. Wang and P. Kollman, J. Comput. Chem., 2003, 24, 1999–2012 CrossRef CAS PubMed.
- U. Essmann, L. Perera, M. L. Berkowitz, T. Darden, H. Lee and L. G. Pedersen, J. Chem. Phys., 1995, 103, 8577–8593 CrossRef CAS.
- A. Jakalian, D. B. Jack and C. I. Bayly, J. Comput. Chem., 2002, 23, 1623–1641 CrossRef CAS PubMed.
- E. Krieger, R. Dunbrack, R. Hooft and B. Krieger, Computational Drug Discovery and Design, 2012, vol. 819, pp. 405–421 Search PubMed.
- R. W. W. Hooft, G. Vriend, C. Sander and E. E. Abola, Nature, 1996, 381, 272 CrossRef CAS PubMed.
- A. Pedretti, L. Villa and G. Vistoli, J. Comput.-Aided Mol. Des., 2004, 18, 167–173 CrossRef CAS PubMed.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Assessment of homology model quality. Considerations upon MD calculations. Validations of MD stability. Validation of the homology model in performing a suitable level of docking accuracy. Movie of ONIOM minimization in avi format. See DOI: 10.1039/c6ra05557e |
| This journal is © The Royal Society of Chemistry 2016 |





